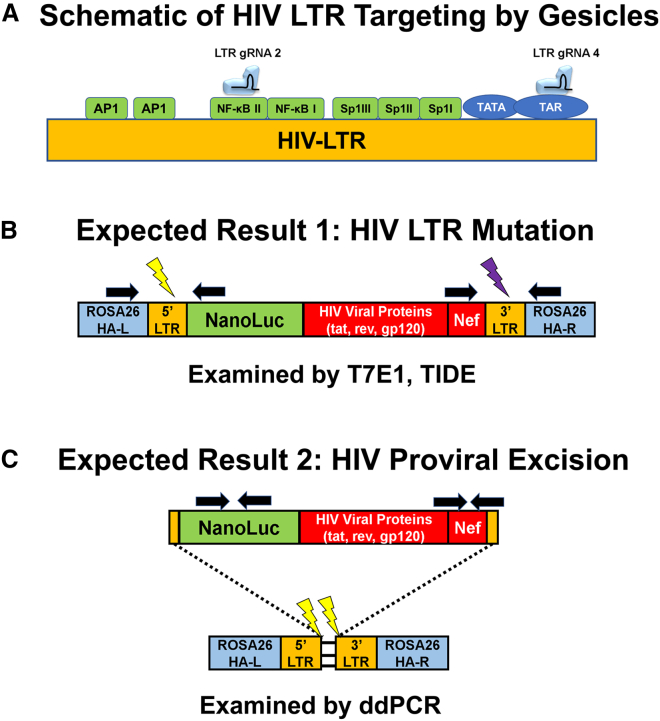
Figure 4.
Schematic of the HIV Long Terminal Repeat and Expected Products after CRISPR/Cas9 Targeting by Gesicles
(A) Two gRNAs were developed to target the HIV LTR. LTR gRNA 2 targeted the NF-κB II transcription site, whereas LTR gRNA 4 targeted the TAR region. (B) The HIV provirus utilizes two LTR regions at the 5′ and 3′ ends. If CRISPR/Cas9 occurs at separate instances (denoted by the light and dark lightning bolts), mutations in these regions can occur. Black arrows denote the primer binding sites to assay mutations within the LTR regions. (C) Additionally, if a double-stranded break occurs at the same instance, there is a possibility of whole provirus excision resulting in a loss of HIV proviral copies. Black arrows denote primers used to assay the loss of integrated proviruses by ddPCR.