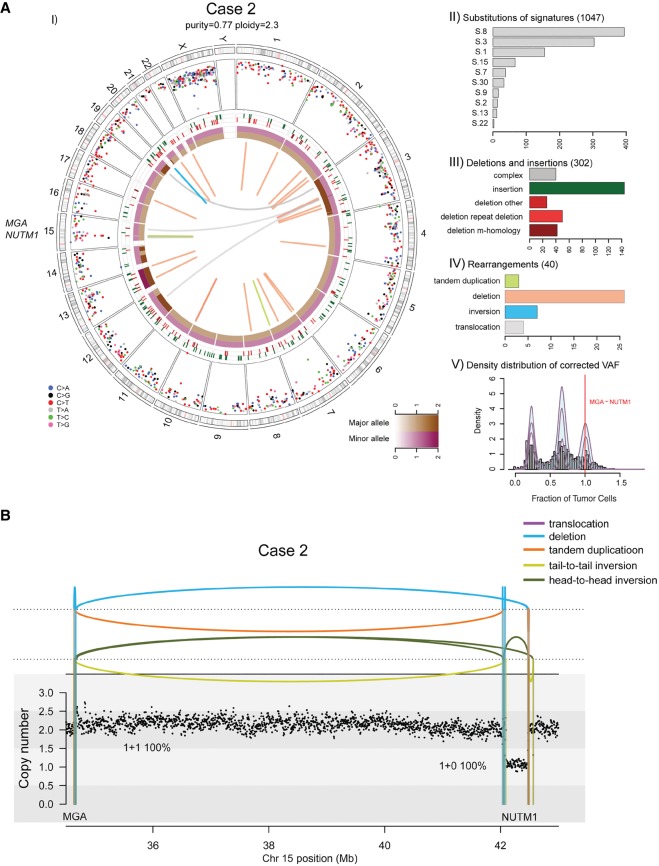
Figure 2.
(A) Whole-genome analysis identifies a novel MGA–NUTM1 fusion in Case 2. (I) Circos plot summarizing the whole-genome sequencing (WGS) data. (II) Mutation signature analysis of the substitutions using Mutational Patterns. (III) Summary of the indel data also showing the contribution of repeat- or microhomology-mediated mechanisms among deletions. (IV) Summary of rearrangement data. (V) Statistical analysis of the corrected variant allele frequency of substitution by Bayesian Dirichlet process–based clustering. For details, see legend to Figure 1. (B) Integrated copy-number/rearrangement plots showing the WGS-based absolute copy number (y-axis) across the indicated genomic region (x-axis). Rearrangements are depicted as lines perpendicular to the x-axis.