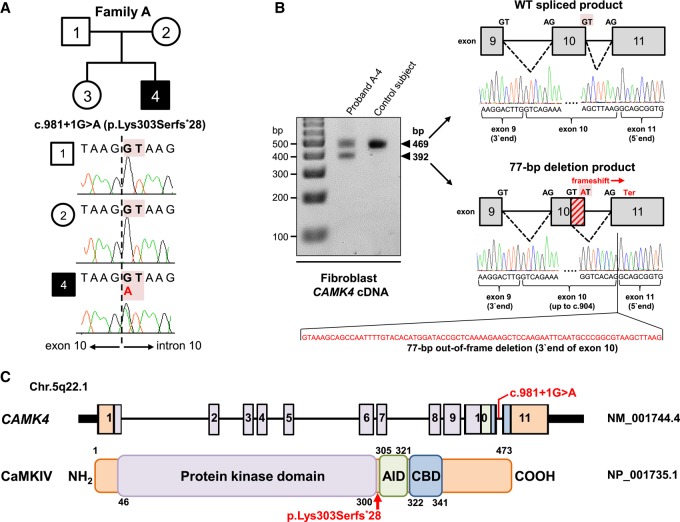
Figure 2.
Identification of a de novo heterozygous essential splice-site variant, c.981+1G>A, in CAMK4 and its predicted consequences at the transcript and protein level. (A) Pedigree for the investigated family A and CAMK4 splice-disrupting variant in genomic DNA. The affected individual (proband A-4) is shaded black. Sanger sequencing confirmed the de novo status of the exome-identified c.981+1G>A variant located within the canonical GT splice donor of the penultimate exon 10 of CAMK4. (B) RT-PCR followed by gel electrophoresis and Sanger analysis in dermal fibroblasts from proband A-4 and a control subject. Apart from the 469-bp normal size cDNA product, proband A-4's cells produce, in equal amount, a second product of higher mobility (392 bp). Direct sequencing revealed that the 392-bp product is the result of utilization of a cryptic GT splice site upstream of the mutated donor, with generation of an abnormal mRNA isoform lacking the final 77 nt of exon 10 (diagonally hashed rectangle), leading to a frameshift and the introduction of a premature termination codon in the last exon of CAMK4 (p.Lys303Serfs*28). WT, wild-type. (C) Diagram (not drawn to exact scale) of the human CAMK4 locus (NM_001744.4) and its encoded protein (NP_001735.1). The identified c.981+1G>A variant is shown relative to exonic structure and the position of the protein truncation (p.Lys303Serfs*28) is highlighted with a red arrow. Numbering refers to exons (upper panel) and amino acids (lower panel). Key protein domains are illustrated in color: purple for the protein kinase domain, green for the AID, and blue for the CBD.