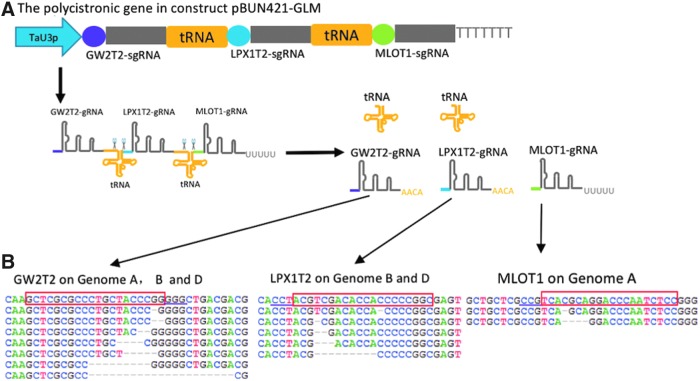
FIG. 1.
CRISPR-Cas9-based multiplex editing in hexaploid wheat using gRNAs processed through the endogenous tRNA-processing system. (A) Schematic of tRNA-based processing of a polycistronic gene transcript. The polycistronic gene containing three tRNA–gRNA blocks was driven by a TaU3 promoter in a MGE construct (pBUN421-GLM). Guide sequences GW2T2, LPX1T2, and MLOT1 are shown in purple, blue, and green, respectively. The GW2T2-gRNA, LPX1T2-gRNA, and MLOT1-gRNA are released after the tRNA processing. (B) The representative next-generation sequencing (NGS) results obtained for three genomic regions targeted by the pBUN421-GLM construct. The wild-type sequences are shown on the top. The target sequences are shown in the red rectangles; the PAM sequences are underlined; the deletions are shown by dashed lines.