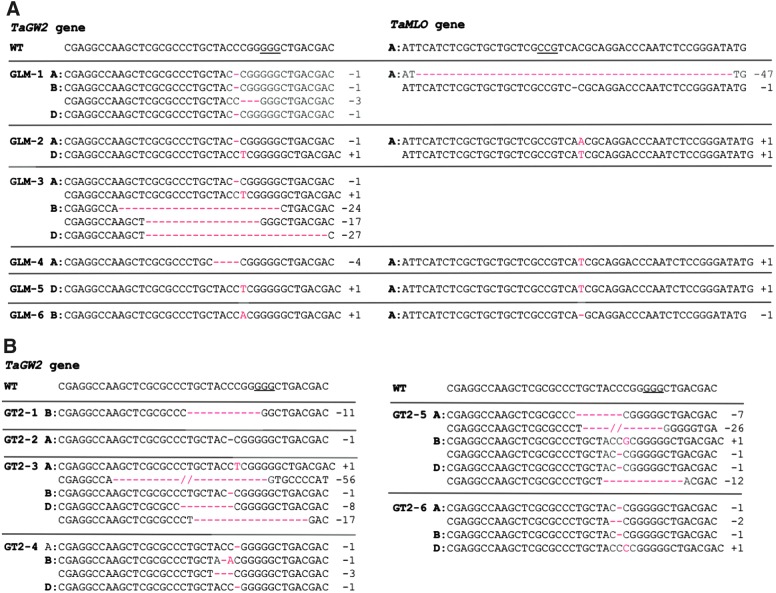
FIG. 2.
CRISPR-Cas9-induced mutations in the T0 transgenic plants identified by NGS. (A) CRISPR-Cas9-induced mutations in the homoeologs of the TaGW2 and TaMLO genes in the T0 plants expressing MGE construct pBUN421-GLM. (B) CRISPR-Cas9-induced mutations in the homoeologs of the TaGW2 gene in the T0 plants expressing single gRNA construct pBUN421-GT2. Only mutated reads with a frequency >30% were shown in (A) and (B) (see Supplementary File 5 for details). WT, wild-type alleles in wheat cultivar Bobwhite; “–” and “+” signs and numbers after them, nucleotides deleted and inserted, respectively. The PAM sequences are underlined; the mutated nucleotides are highlighted in red.