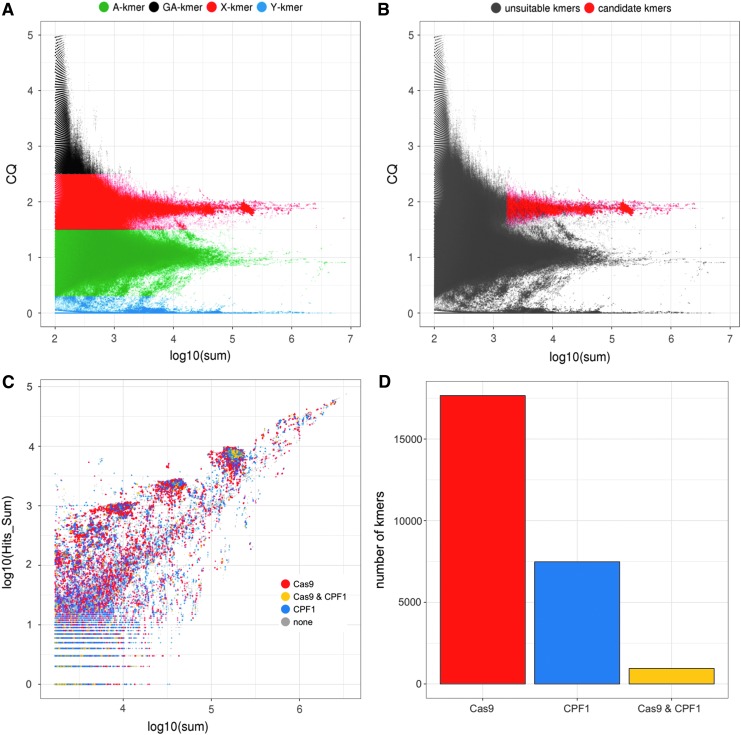
FIG. 4.
kmer analysis and selection in the Pimperena strain. (A) kmer-CQ versus abundance (in log10 of sum) in both male and female Illumina libraries for all 270 million redkmer-generated kmers colored by chromosomal bins. (B) Redkmer selection plot showing kmer-CQ versus abundance for predicted X-chromosome-specific and abundant kmers (red dots) versus the unsuitable kmers (gray dots). (C) Identification of kmers containing sequences predicted to be suitable for targeting by CRISPR endonucleases showing kmer abundance in the Illumina data (x-axis) versus their occurrence within PacBio data (y-axis). Kmers lacking suitable characteristics for CRISPR (e.g., the absence of a PAM motif) are shown as gray dots, kmers suitable for Cas9 in red, kmers suitable for Cpf1 in blue, and those harboring sequences suitable for both Cas9 and Cpf1 are shown in yellow. (D) Number of kmers containing sequences suitable for CRISPR nuclease platforms.