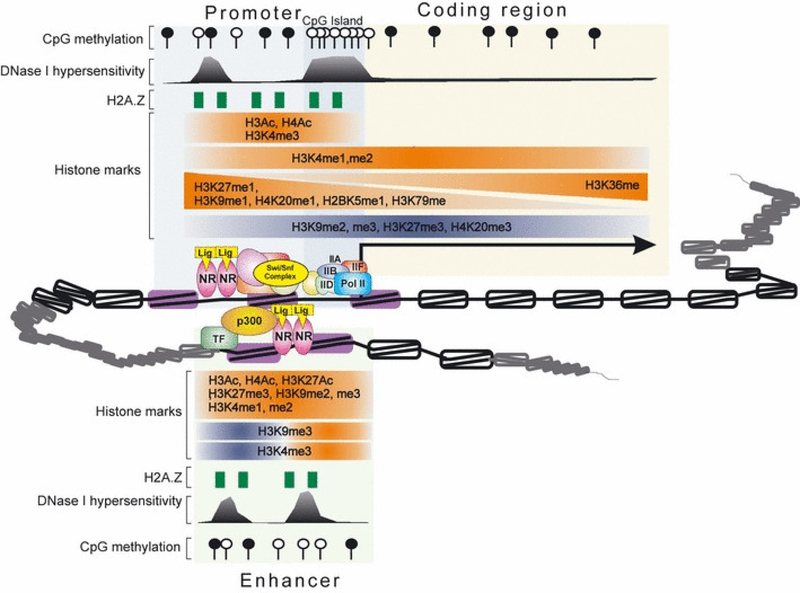
Fig. 3.
Characteristics of local chromatin structures within promoters, enhancers and coding regions. The non-random positioning of a nucleosome is dictated by DNA sequence, activity of remodeling complexes (like SWI/SNF) and competition of the nucleosome with TFs for access to specific DNA sequences. The regulatory regions are characterized by high turnover of histone proteins (depicted by purple nucleosomes). The histone marks identified at the promoters and enhancers of active (red) and silent (blue) genes are indicated. The gradients reflect changes of histone marks across the coding region. Contradictory observations about the presence of H3-K9me3 and H3-K4me3 within enhancer regions have been reported. Both promoters and enhancers are marked by DHSs and H2A.Z histone variants. Most promoters are characterized by increased density of CpG dinucleotides (CpG islands) which are usually unmethylated (open circles). Enhancers also show highly localized CpG enrichment with DNA methylation status correlating with their activity. The CpG dinucleotides are under-represented within coding regions and contain high methylation levels (filled circles) in order to prevent spurious transcription.