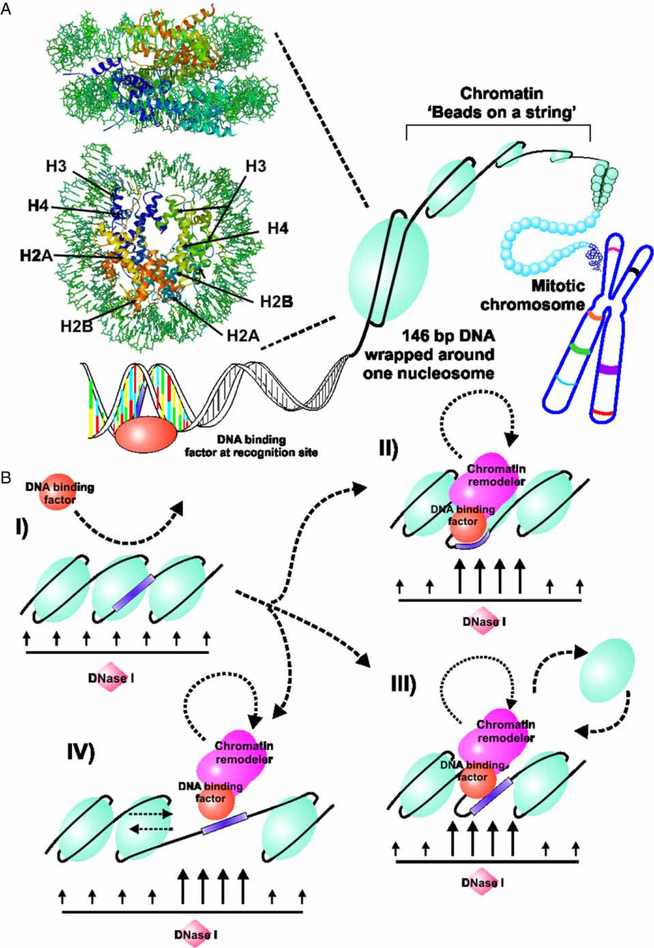
Figure 2.
Interaction of transcription factors with chromatin. (A) DNA in the mammalian cell is organized in a hierarchical series of nucleoprotein structures collectively referred to as chromatin. The basic repeating unit in this structure is the nucleosome, with 146 bp of DNA wrapped on an octameric core of histones (H). The nucleosome is a flattened ellipsoid approximately 110Å on the large axis and 57Å thick. This structure is rendered here in RasMol from the coordinates published for the Xenopus laevis particle by Richmond and colleagues (Ong et al. 2007). Although transcription factors recognize specific sequences in DNA, access to these sites is severely constrained by packaging of the sequences in the nucleosome and higher order structures. (B) Eukaryotic cells have evolved complex systems to reorganize local nucleosome structures during gene regulation. These systems, referred to as remodeling enzymes, are recruited to specific regions by protein–protein interactions with transcription factors, and transiently “open” the nucleosome structure. These molecular reorganizations can be conveniently detected and mapped by the local increase in access to DNA by nucleolytic reagents, including DNaseI (Wu 1980), and chemicals such as methidiumpropyl-EDTA-Fe(II) (Cartwright and Elgin 1989). Multiple mechanisms are considered to be involved in nucleosome remodeling. A pioneer protein, such as GR, can sometimes recognize sites in non-remodeled chromatin (I), and initiate a cascade of binding events. These events can include the transient remodeling of core histone structure (II), the complete or partial eviction of histone components (III), or the “sliding” of nucleosomes to new translational positions on the DNA (IV). The mobility of transcription factors on DNA is now understood to be linked to these chromatin remodeling processes.