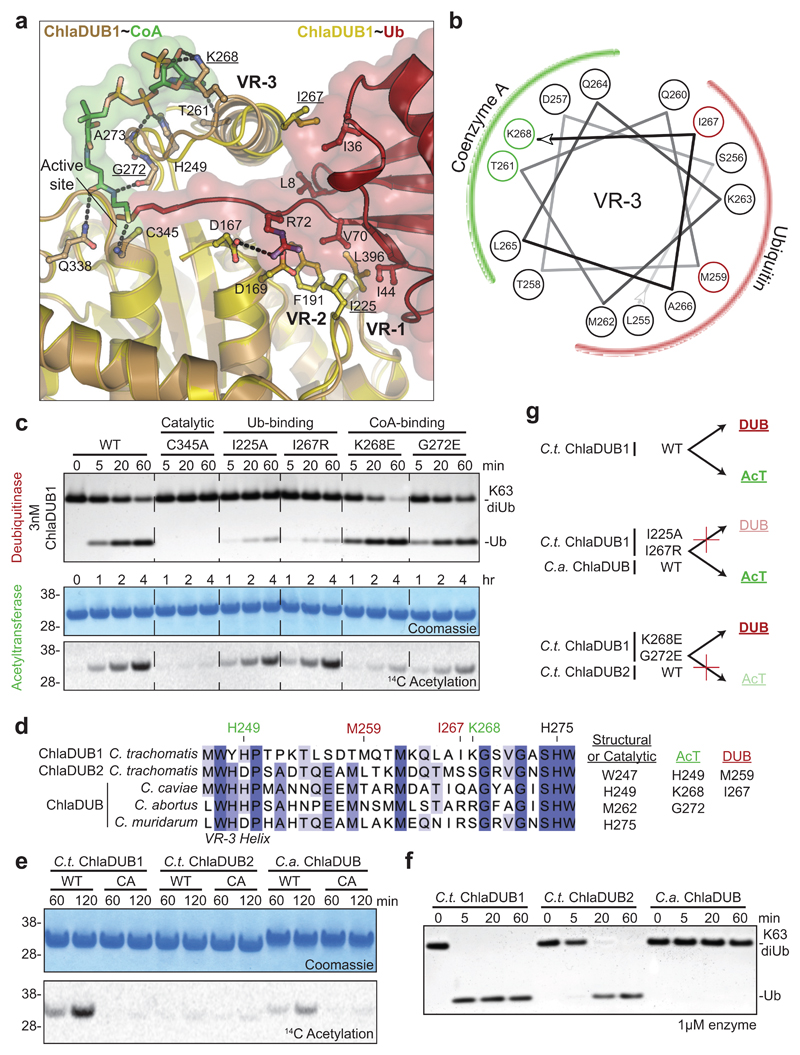
Figure 2. Molecular dissection of dual deubiquitinase/acetyltransferase activities.
a) Close-up of the ChlaDUB1:CoA (brown:green) and ChlaDUB1:Ub (tan:red) interfaces with key interacting residues shown in ball-and-stick. Hydrogen bonds are shown as dashed lines. b) Helical wheel diagram illustrating the amphipathic nature of the ChlaDUB1 VR-3 helix, and its interactions with Ub or CoA (colored in red and green, respectively). c) Deubiquitinase (top) and acetyltransferase (bottom) assays illustrating that while both activities require the catalytic Cys residue, mutations in the Ub-binding and CoA-binding regions separate the two functions. A representative gel is shown of triplicate experiments. d) Sequence alignment of the VR-3 helix for orthologous Chlamydia ChlaDUB enzymes. The catalytic His, CoA-binding (green) and Ub-binding (red) residues are marked, and additional contacts are listed. e) 14C acetylation assay with the ChlaDUB orthologues from C. trachomatis (C.t. ChlaDUB2) and C. abortus (C.a. ChlaDUB). f) Deubiquitinase assay monitoring K63-linked diUb cleavage by the Chlamydia ChlaDUB orthologues. Gels in e and f are representative of triplicate experiments. All uncropped gels are shown in Supplementary Fig. 10. g) Schematic depicting how deubiquitinase and acetyltransferase functions can be separated either by structure-guided mutation or evolution as represented by the ChlaDUB orthologues.