Figure 1.
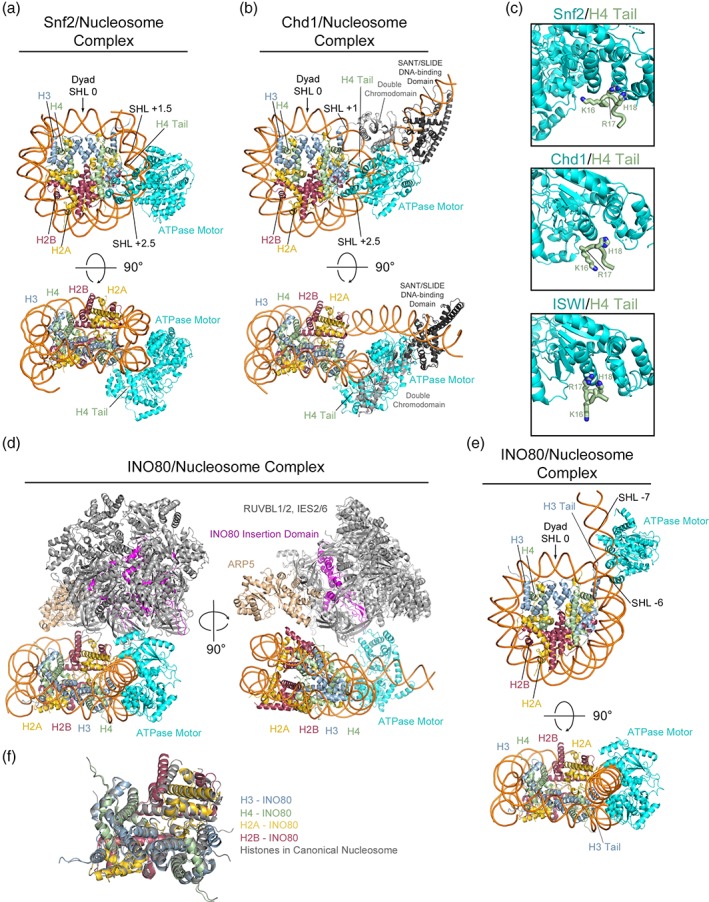
Cryo‐EM structures of Snf2, Chd1, and INO80 chromatin remodelers bound to nucleosomes. (a) Front and top view of 4.69 Å structure for S. cerevisiae Snf2 bound to a nucleosome, details of the interactions with nucleosomal DNA and the H4 tail are highlighted (PDB: 5X0Y). (b) Front and top view of 4.80 Å structure for S. cerevisiae Chd1 bound to a nucleosome, details of the interactions with nucleosomal DNA and the H4 tail are highlighted (PDB: 5O9G). (c) Comparison of the interaction between H4 N‐terminal tail residues 15–18 and the ATPase motor domains from Snf2 (top), Chd1 (middle), and ISWI (bottom) (PDB: 5JXT). (d) 4.8 Å structure of the human INO80 complex bound to a nucleosome (PDB: 6ETX). A rotated view details the assembly of the RUVBL1/2 heterohexamer and additional subunits on the INO80 insertion domain and highlights the locations of the INO80 ATPase motor domain and ARP5 subunit. (e) Front and top view of the INO80 ATPase motor bound to a nucleosome with additional domains and subunits removed for simplicity. Details of the interactions with nucleosomal DNA and the H3 tail are highlighted. (f) Alignment of the histone octamer from the INO80 complex structure (PDB: 6ETX) with the histone octamer from the crystal structure of the nucleosome alone (PDB: 1AOI) detailing the rearrangement on INO80 binding.
