Figure 2.
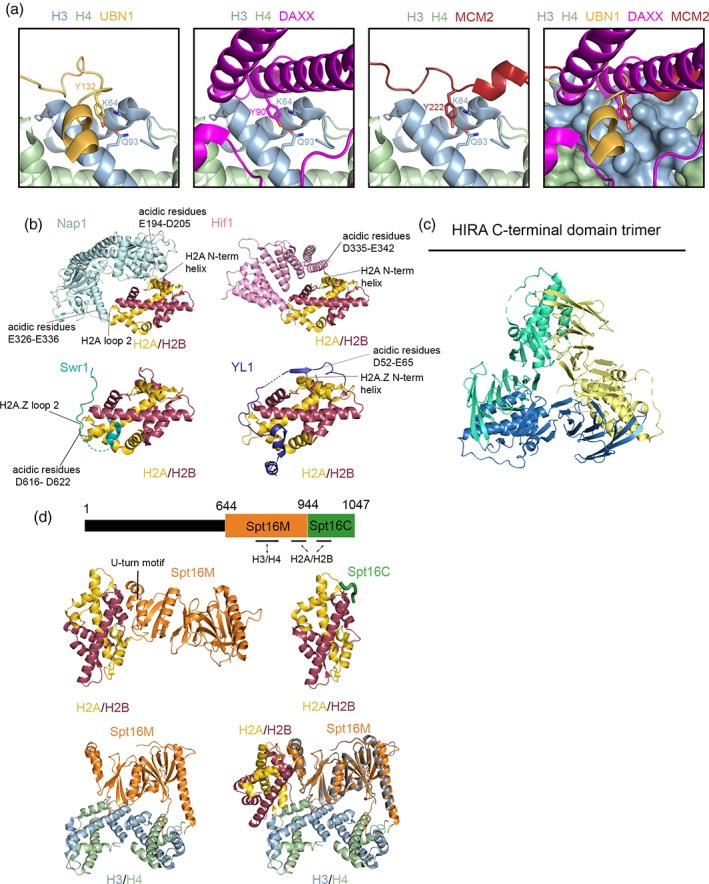
Molecular comparison of different histone chaperone proteins. (a) UBN1 (PDB: 4ZBJ), DAXX (PDB: 4HGA), and MCM2 (PDB: 5BNV) in complex with H3/H4. UBN1 Y132, MCM2 Y90, or DAXX Y222 fit into a surface pocket on H3 where it forms a hydrogen bond with H3Q93 and van der Waals interactions with H3K64. This deep association likely drives competition for H3/H4 binding to ensure orderly histone trafficking in a crowded cellular environment. (b) At least four uniquely functioning H2A/H2B chaperones, Nap1 (PDB: 5G2E), Hif1 (PDB: 5BT1), YL1 (PDB: 5FUG), and Swr1 (PDB: 4M6B), utilize patches of acidic residues to interact with the same basic residues on the negatively charged DNA‐binding surface of an H2A/H2B dimer to sequester the histones from DNA until the final step of nucleosome formation. (c) 2.45 Å crystal structure of the C‐terminal domain of HIRA (residues 644–1017; PDB: 5YJE) reveals homotrimer formation. (d) Top: Spt16 with identified H2A/H2B‐ and H3/H4‐binding domains. Middle: 2.35 Å resolution structure of Spt16M in complex with H2A/H2B (PDB: 4KHA) versus 1.8 Å resolution structure of Spt16C in complex with H2A/H2B (PDB: 4WNN) reveals that these two domains compete for some of the same H2B residues and may represent two unique interactions with H2A/H2B relevant to the FACT mechanism. Bottom: 2.98 Å resolution structure of Spt16M in complex with an H3/H4 tetramer (left; PDB: 4Z2M) overlayed with the Spt16M/H2A/H2B structure (right; H2A/H2B‐bound Spt16M in gray) demonstrates an additional, distinct histone binding domain associated with H3/H4, a feature potentially useful in H3/H4 tetramer stabilization and H2A/H2B dimer shuttling during chromatin reorganization.
