Figure 3.
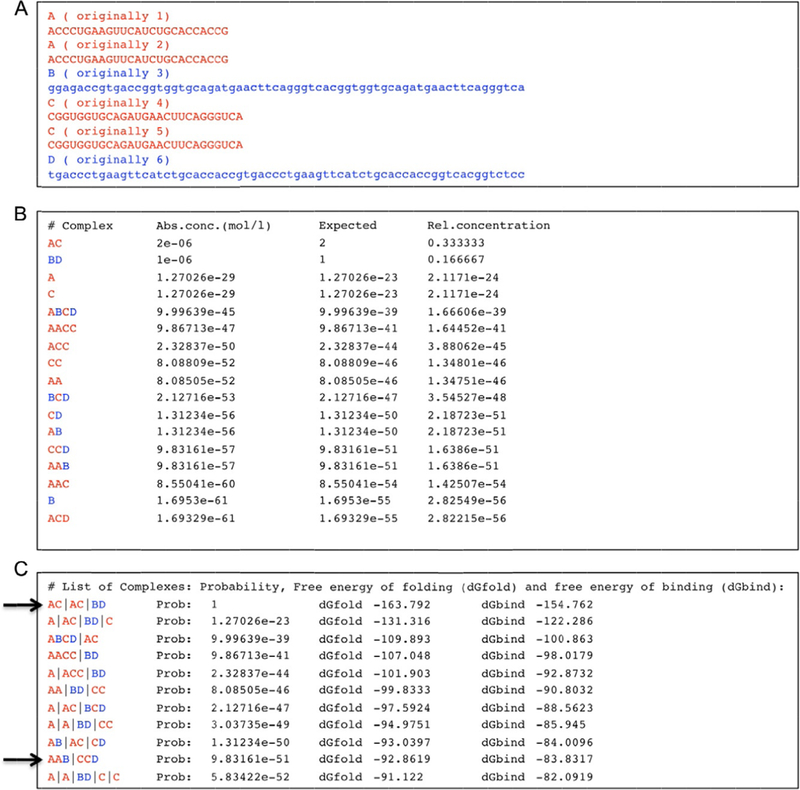
Example of program output for given dual-release RNA (in red; light gray in the print version)/DNA (in blue; dark gray in the print version) hybrid sequences. (A) Input consisting of two copies of sense DS RNAs, two copies of antisense DS RNAs, and two different cognate DNA strands with toeholds. The program notices that sequences 1 and 2 as well as 4 and 5 are identical, and renames them as “A” and “C,” respectively. (B) Part of the output of the program. The predicted complex formation includes DS RNAs (named by the program “AC”) as well as DNA duplexes (named “BD”). Note the formation of complexes named AAB and CCD that indicate the formation of dual-release RNA/DNA hybrids. The computer output shows predictions for the absolute concentrations (“Abs conc.”), the expected number of complexes in the simulation volume and the relative concentration (the expected number of complexes divided by the number of simulated strands). (C) Another list that is part of the output shows secondary structures and their probabilities (“Probability”), free energies of folding (dGfold), and free energies of binding (dGbind). The computer results are based on a list of stems with a minimum length of 3 base pairs; only stems that cannot be extended further are considered. There are no restrictions in terms of pseudoknot complexity. For clarity, symbols A and C indicating RNA strands have been colored red (light gray in the print version), and symbols B and D (indicating DNA strands) have been colored blue (dark gray in the print version); tab characters have been inserted into the computer output. The arrows indicate the desired hybrid state and product states. One possible explanation for the estimated low probabilities of higher order complexes could be the simplification that only secondary structure states are considered in which all helices consist of at least 3 base pairs. Also, the theoretical treatment does not consider the exchange of molecules with the environment.
