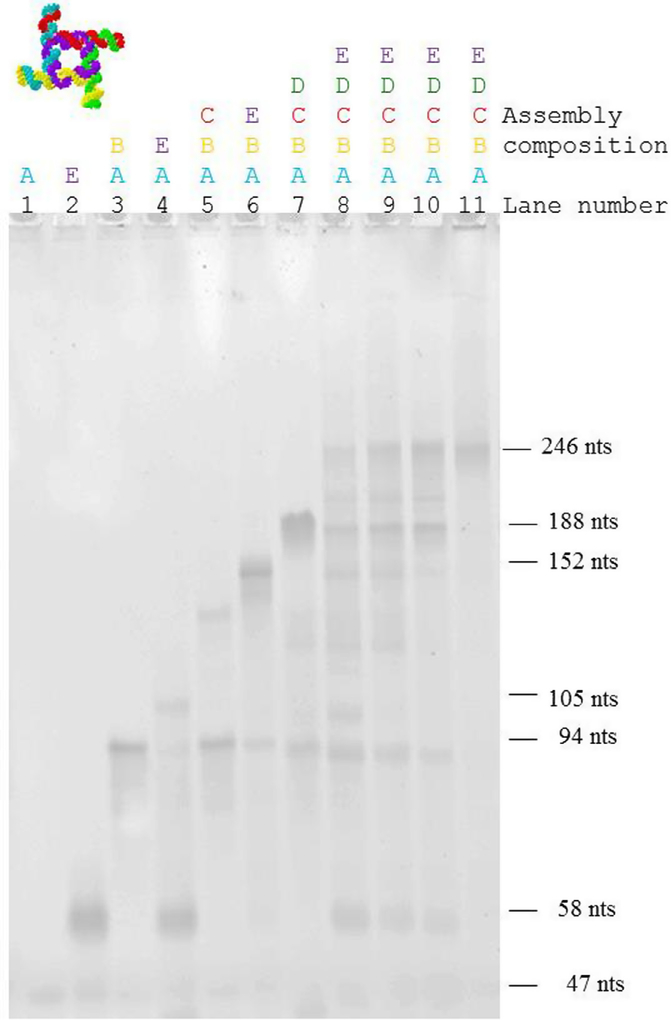
Fig. 6.
Formation of molecular assemblies of the square nanoconstruct. Monomers (lane 1: strand A, 47 nts; lane 2: strand E, 58 nucleotides), dimers (lane 3: strands A + B, 94 nts; lane 4: strands A + E, 105 nts), trimers (lane 5: strands A + B + C; lane 6: strands A + B + E), tetramer (lane 7: strands A + B + C + D) and pentamers (strands: A + B + C + D + E, lanes 8–11). As expected, the dimer A + E (corresponding to 105 nts) displays a slower electrophoretic mobility compared with the dimer A + B (corresponding to 94 nucleotides). Only fractions of strands A and E assemble in a trimer, and the prominent band for the monomer E is observed for this assembly. The trimers formed by strands A, B, and C, and A, B, and E assemble more readily. This can be explained by the difference in the interaction strength between two strands in the dimer, and three strands in the trimer. The tetramer (strands A, B, C, and D,) seems to be thermodynamically less stable, and bands corresponding to the monomer, dimer, trimer and tetramer can be observed for the assembly. Lanes 1–10 correspond to 2 mM Mg++ concentration, while lane 11 corresponds to 4 mM Mg++ concentration. Lanes 8–11 correspond to the pentamer (strands A,B,C,D,E) with different assembly conditions (lane 8: 37 °C, lane 9: 45 °C, lane 10: 55 °C (2 mM Mg), lane 11: 55 °C (4 mM Mg)). The pentamer assembles as expected. At 55 °C and 4 mM Mg concentration, the square assembles without formation of lower molecular weight assemblies. The color-coding corresponds to the molecular models shown in Figs. 5b and e.