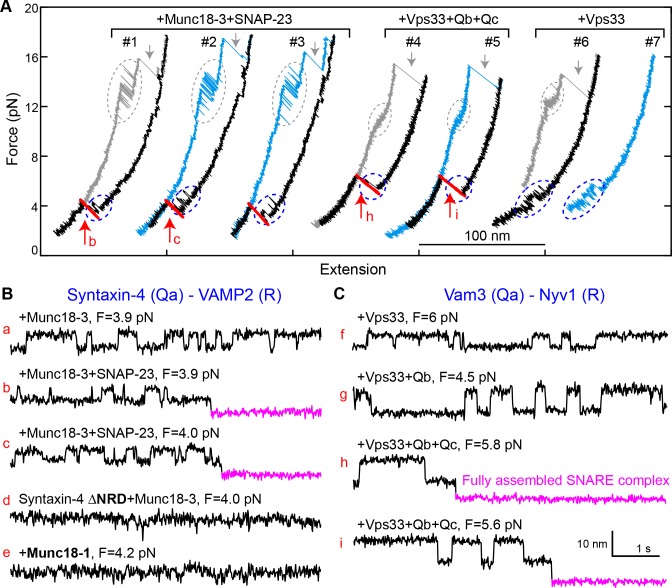
Figure 10. Munc18-3 and Vps33 catalyze SNARE assembly via template complexes.
(A) FECs of the Munc18-3 or Vps33 cognate Qa-R SNARE conjugate in the presence of the indicated protein(s). See also Figure 10—source datas 1 and 2. (B–C) Extension-time trajectories at the indicated constant mean forces, some of which (b, c, h, and i) are extracted from panel A. See also Figure 10—source datas 3 and 4.
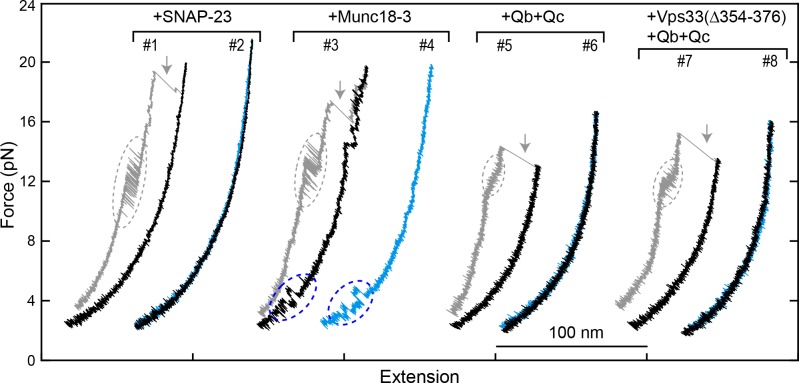
Figure 10—figure supplement 1. FECs obtained by pulling and relaxing a single syntaxin-4-VAMP2 conjugate (#1–4) or Vam3-Nyv1 conjugate (#5–8) in the presence of the indicated protein or proteins.
Figure 10—figure supplement 2. FECs displaying Vps33-catalyzed vacuolar SNARE assembly, marked by red arrows.
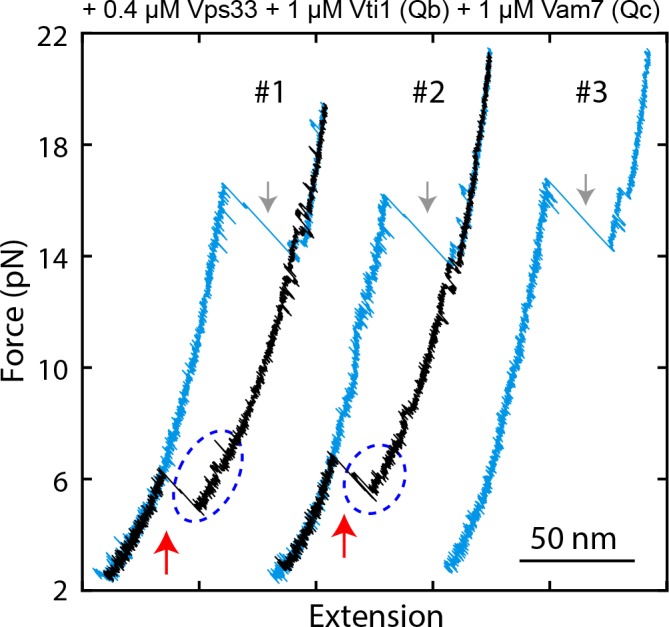
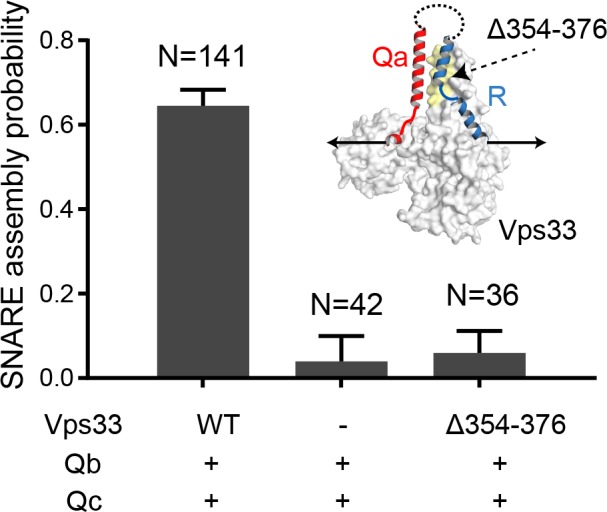
Figure 10—figure supplement 3. Probabilities of vacuolar SNARE assembly per relaxation under different conditions.