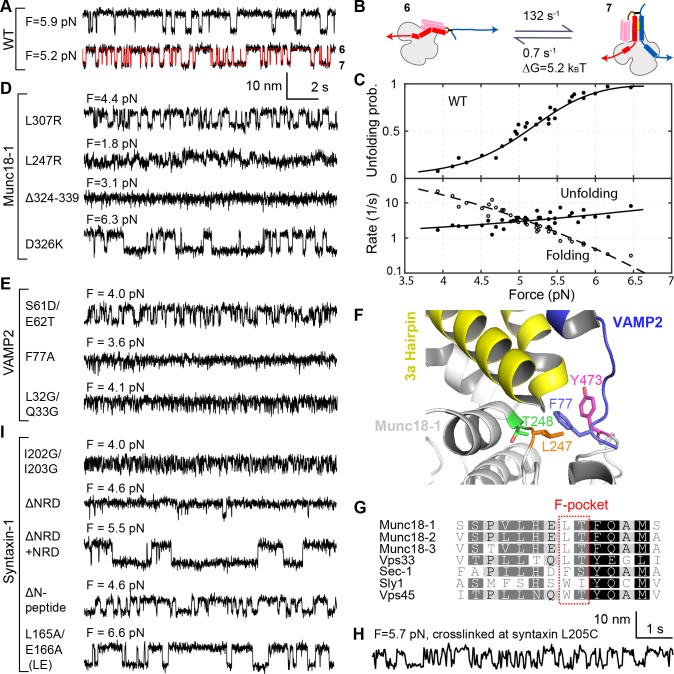
Figure 3. Stability, conformation, and folding kinetics of the template complex.
(A, D, E, I) Extension-time trajectories at constant mean forces with the WT template complex (A) or its variants containing indicated mutations in Munc18-1 (D), VAMP2 (E), or syntaxin (I). The red trace in A shows an exemplary idealized trajectory derived from hidden Markov modeling. Trajectories in A, D, E, and I share the same scale bars. See also Figure 3—figure supplement 1 and Figure 3—source datas 1 and 2 in Dataset 1. (B) Diagram illustrating the transition between the partially closed syntaxin state (state 6 in Figure 2D) and the template complex state (state 7); rates and energies are derived from panel C. (C) Force-dependent unfolding probabilities (top) and transition rates (bottom). Best model fits (solid and dashed curves) reveal the stability and folding and unfolding rates of the template complex at zero force (Figure 4, Table 1, and Figure 3—source data 3 in Dataset 1). (F) Structural model of VAMP2 F77 anchored in the F-pocket in Munc18-1 composed of L247 and T248, which is covered by Y473. The model was derived by superimposing the structures of Munc18-1:syntaxin (Figure 2B; 3C98) and Vps33:Nyv1 (5BV0). (G) Sequence alignment showing F-pocket sequence conservation among SM proteins. (H) Extension-time trajectory of the WT template complex at 5.7 pN. The Qa-R SNAREs were crosslinked between syntaxin L205C and VAMP2 Q36C (Figure 2A, open arrowhead). See also Figure 2—figure supplement 1 and Figure 3—source data 2 in Dataset 1.