Extended Data Fig. 4 |. Detection of TET2 chimaeric transcripts in Patient-10 CAR T cells and DNA sequencing for mutation detection.
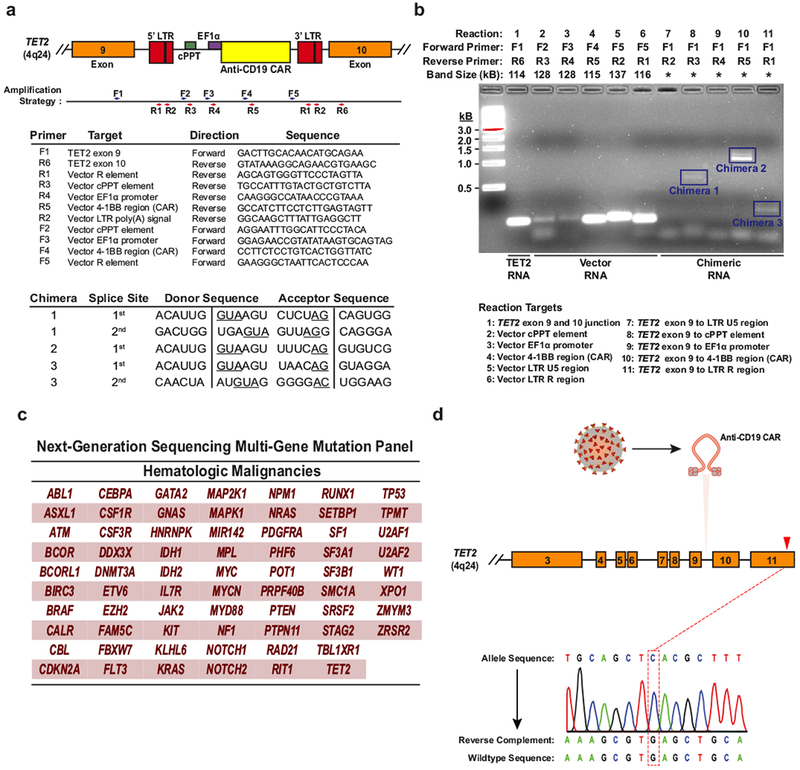
a, The strategy for detection of polyadenylated RNA corresponding to truncated TET2 transcripts is depicted. Boxes represent the genomic regions between TET2 exons 9 and 10 with the integrated vector present. Blue and red arrows indicate general locations of the forward and reverse primers, which are listed below the diagram. LTR, long terminal repeat; cPPT, polypurine tract; EF1α, elongation factor 1-α promoter. Sequences corresponding to the splice junctions for the three chimaeric messages (five total junctions) are listed in the bottom chart. Underlines indicate consensus splice donors and acceptors. b, Visualization of chimaeric TET2 RT–PCR products. PCR products were separated on a native agarose gel and stained with ethidium bromide. Expected sizes of amplicons are listed above the gel. Truncated transcripts are highlighted by blue boxes. A key to the RT–PCR reactions is shown below the diagram. *Band size not determined (two independent experiments). c, Genes interrogated by the next generation sequencing panel used to analyse DNA isolated from CD8+CAR+ T cells and CAR− T cells in Patient-10 at the peak of his response. d, Sanger sequencing of specific amplifications corresponding to the allele that was disrupted by integration of the CAR lentivirus is shown. The mutation that was detected by next generation sequencing of total genomic DNA from CAR+ T cells (Fig. 3c) is not present in the TET2 allele hosting the lentiviral integration site.
