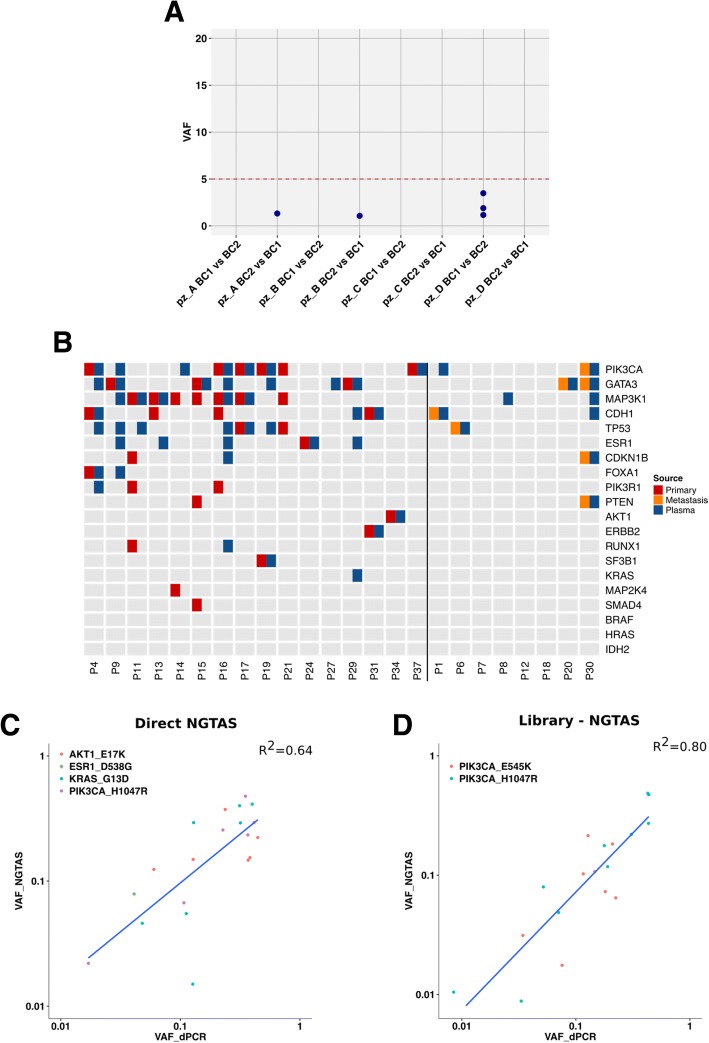
Fig. 5.
Validation of NG-TAS performance in clinical plasma samples. a The specificity of NG-TAS in clinical samples was estimated using 4 pairs of buffy coats from the same patients (A, B, C and D). The mutation calling pipeline was applied using one buffy coat as normal and the other as ‘tumour’ and vice versa. All mutations called in this setting can be considered FPs. The red line indicates 5% VAF. b Oncoprint summary plot of genes mutated in 24 cases for which both tissue and plasma samples were tested. The vertical black line separates cases for which the primary tumour was analysed from cases for which a metastasis biopsy was analysed. c, d Comparison of VAF obtained by NG-TAS and dPCR. c In this comparison, four different hotspot mutations including AKT1 (E17K), ESR1 (D538G), KRAS (G13D) and PIK3CA (H1047R) identified in multiple plasma samples from 4 distinct patients were analysed (R2 = 0.64). d Two PIK3CA hotspots (H1047R and E545K) were detected by NG-TAS using NGS library as an input material in plasma samples from two distinct patients. The same mutations were detected using dPCR, and a good correlation was found (R2 = 0.80)