Figure 3.
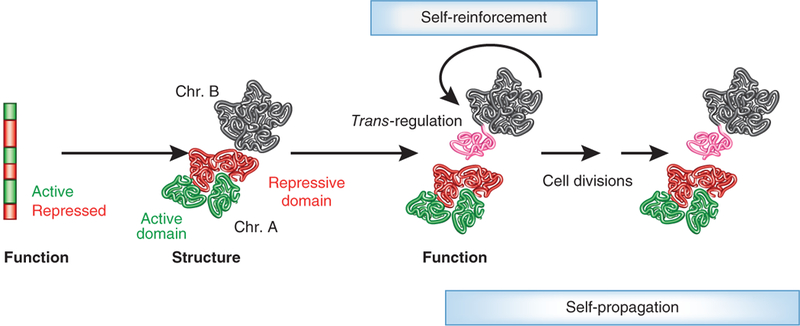
A model depicting the interplay of genome structure and function. The transcriptional activity of genome regions determines the formation of chromatin domains (red and green). Domains are defined patterns of nucleosome positioning, histone modifications and differential higher-order folding. The activity state of a ‘neutral’ genome region (black) is determined by its physical association with either an active or repressive environment, and these long-range contacts may thus change functional states (indicated by transformation of the portion of the black chromosome closest to a repressive (red) domain of another chromosome to pink). The functional status of the chromatin domain feeds back and reinforces its structural features (self-enforcement). Chromatin structure-function relationships are heritable (self-propagation).However, given the inherent plasticity of the system, even in terminally differentiated states strong physiological or environmental stimuli may switch chromatin domains, allowing for the possibility of cell reprogramming.
