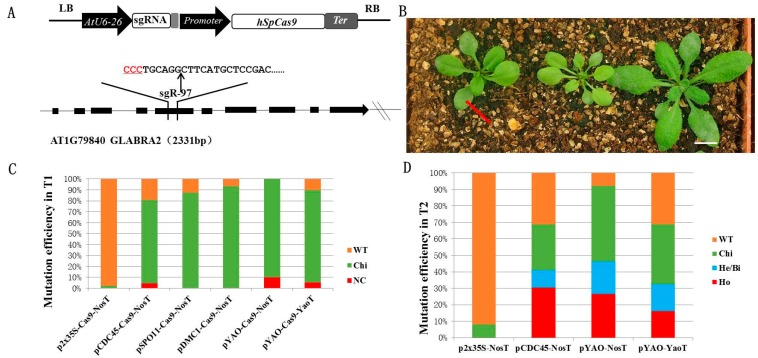
Figure 1.
Gene mutagenesis efficiency of cell division specific CRISPR/Cas9 systems in transgenic T1 and T2 generations of Arabidopsis. (A) Structure of the CRISPR/Cas9 binary vectors and sequence of the target site in GLABRA2. The hSpCas9 gene is driven by the cell division specific promoters CDC45, DMC1, SOP11, and YAO as well as the constitutive 2x35S CaMV promoter, while the AtU6-26 promoter controls transcription of the sgRNA. The sequence of GL2-sgR97 is shown. hSpCas9, human codon-optimized SpCas9; Ter, Nos or YaoT terminator. The PAM sequence is underlined in red. (B) Phenotypes of T1 generation plants transformed with the pCDC45-Cas9 construct. A chimera showing an incomplete glabrous phenotype, a plant showing a uniform glabrous phenotype and a wild type phenotype are shown from left to right. T1 seedlings were transplanted to soil and grown under long-days (16 h light/8 h dark) for two weeks before photographing. Red arrow shows a leaf with a chimeric phenotype. Bar: 1 cm. (C) Mutation efficiency in transgenic T1 lines. Approximately 40 individual T1 plants were tested for each construct. NC: non-chimeric mutants (homozygous, bi-allelic, heterozygous); Chi: chimera; WT: wild type. (D) Mutation efficiency in transgenic T2 generation. Ho: homozygous; He/Bi: heterozygous/bi-allelic; Chi: chimera; WT: wild type.