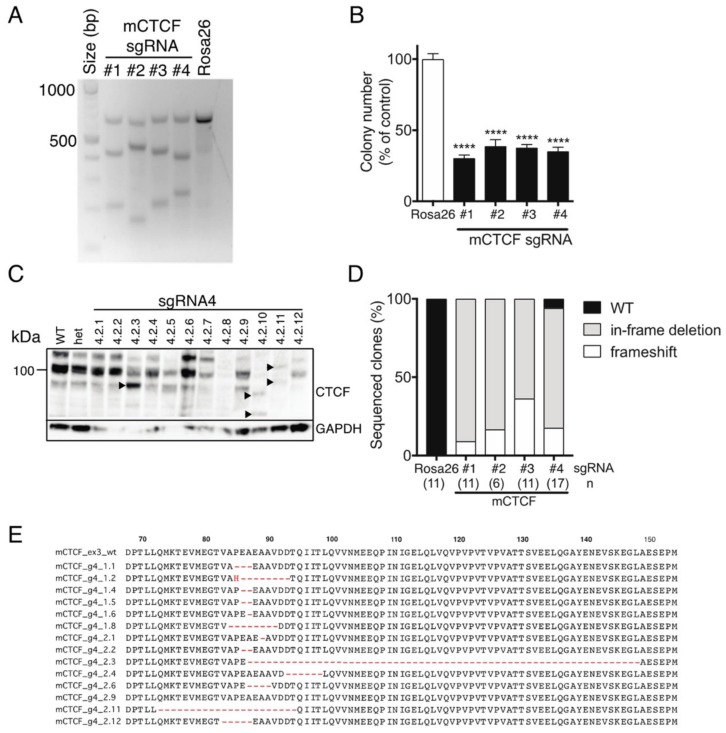
Figure 5.
CRISPR/Cas9-directed editing of Ctcf in hemizygous MEFs. (A) Ctcf+/pgkneo MEFs transduced with Cas9 and sgRNA-containing lentivectors (mouse Ctcf exon 3 sgRNAs #1, #2, #3, and #4; Rosa26 sgRNA) were FACS-enriched and subjected to T7EI digestion of Ctcf exon 3 amplicons amplified from isolated gDNA. Approximate expected sizes (in bp) for digested products #1 (427, 214), #2 (476, 165), #3 (428, 213), and #4 (399, 242). Clonogenicity assay (B); Western blot analysis of individual clones (from sgRNA#4, arrowheads indicate lower molecular weight Ctcf variants) (C); and molecular genetic analysis of individual clones; n=number of clones sequenced (in brackets) (D). (E) Examples of frequently occurring in-frame deletions in Ctcf+/pgkneo MEFs (from sgRNA#4). Quantitative data represent the mean ± SEM for three experiments each performed in triplicate. Statistical analysis was performed using Mann-Whitney U-test (ns = not significant, **** p < 0.0001).