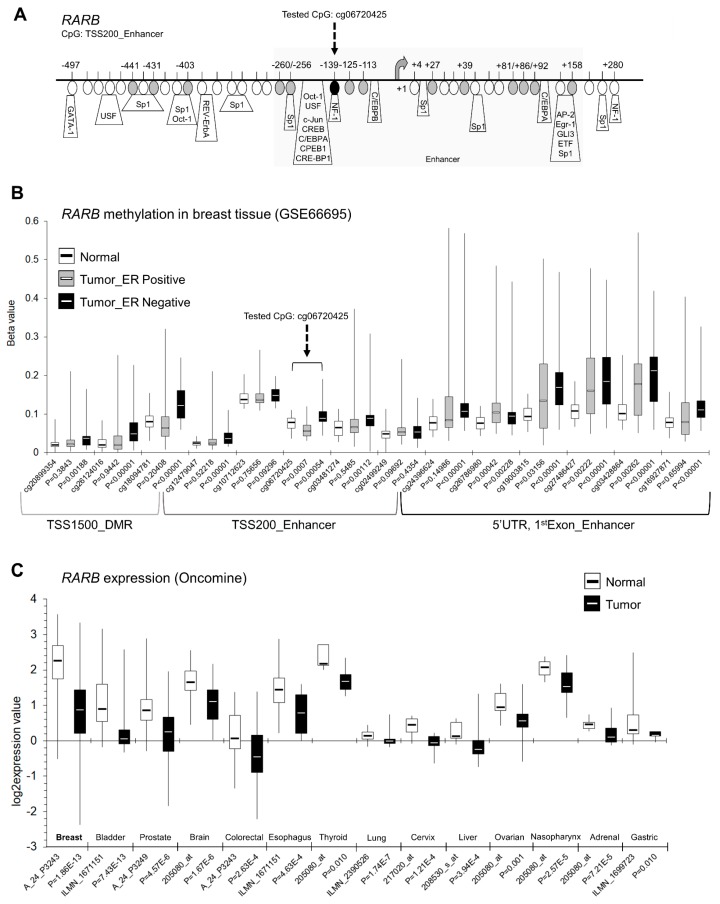
Figure 3.
Relevance of DNA methylation-mediated silencing of RARB tumor suppressor in human breast cancer in vivo. (A) A map of the RARB proximal promoter region (Human GRCh37/hg19 Assembly) including an enhancer region. The methylation-sensitive restriction analysis (MSRA)-tested CpG site (located within the enhancer region of the RARB promoter, -139 bp from transcription start site (TSS); cg06720425 on Illumina 450K microarray platform; chr3: 25469694) is depicted as a black circle, indicated with black dashed arrow. The neighboring CpG sites [-441 bp to +158 bp from TSS] covered on Illumina 450K array are depicted as gray circles. Putative transcription factor binding sites are indicated as predicted using TransFac. (B) Methylation status of these 14 CpG sites (black and gray circles on the gene map) within the RARB proximal promoter region, covered on Illumina 450K array and expressed as beta value in breast tumors based on publicly available Illumina 450K data (GSE66695). The tested CpG site, cg06720425, is indicated with black dashed arrow. Beta value, the methylation score for a specific CpG site with any values between 0 (unmethylated) and 1 (completely methylated), according to the fluorescent intensity ratio. (C) Breast cancer gene expression microarray data for RARB from Oncomine. The normal versus tumor gene expression data are presented as log2-transformed median centered per array, and SD-normalized to 1 per array. The demonstrated changes are statistically significant (P < 0.05).