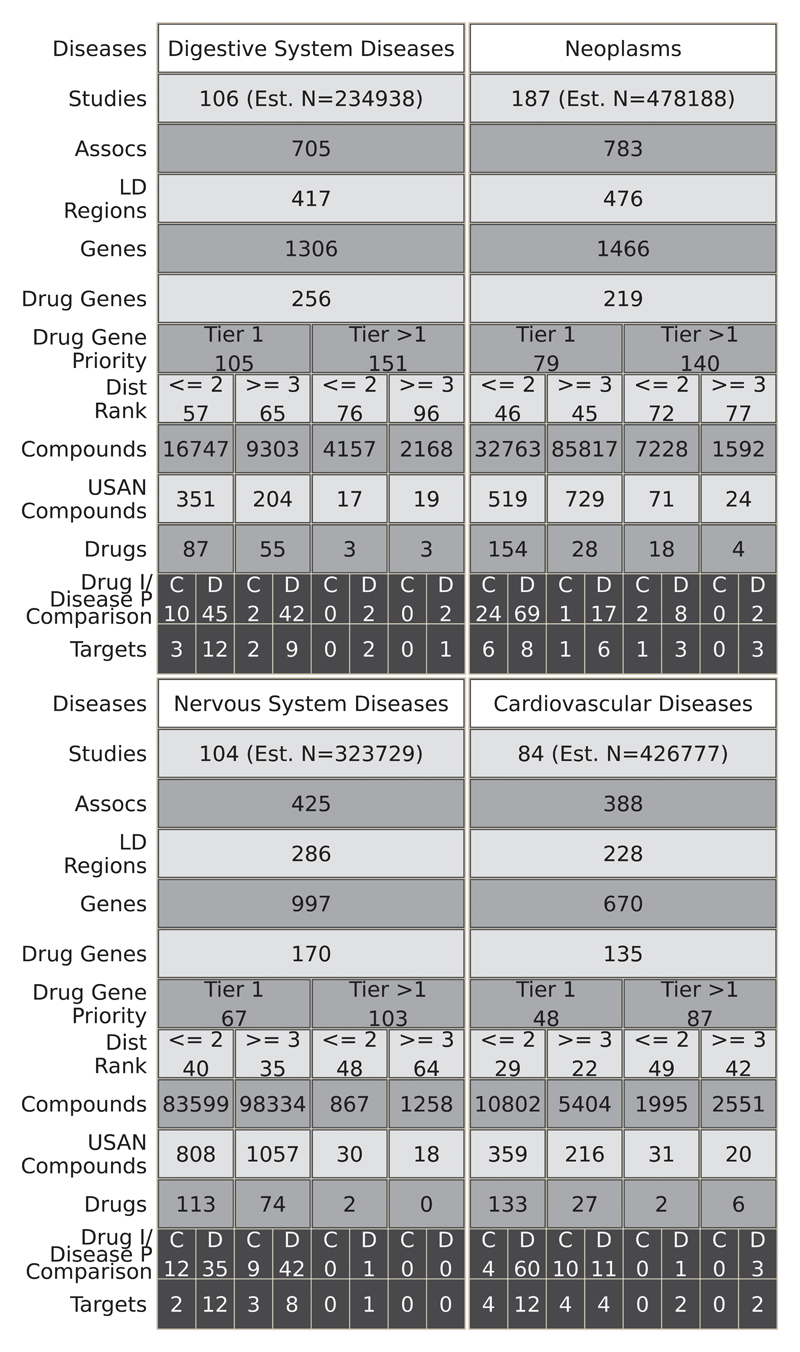
Fig. 5.
Translational potential for the top 4 most studied MeSH root disease areas. For each disease area, the figure illustrates the estimated number of GWAS (Studies Row), the number of associations (p≤5×10-8) (Assocs), the number of LD regions corresponding to those associations (LD Regions), the number of genes in those regions (Genes), and the number of those genes that encode druggable targets (Drug Genes). Subsequent rows quantify the number of druggable genes by priority tier (Drug Gene Priority) and by distance rank of the druggable gene from the GWAS SNP (Dist Rank). The total numbers of compounds (Compounds), compounds with an USAN/INN (USAN Compounds), and drugs corresponding to the drugged targets are also listed (Drugs). In the penultimate row, the numbers of drugs with an indication that is concordant (C) or discordant (D) with the GWAS phenotype are displayed (Drug I/Disease P Comparison). In the final row, the numbers of cognate targets for the concordant or discordant drugs are shown (Targets). Note that for the purposes of the figure, a drug target is a single gene even if it is part of a complex that is targeted by the drug. Within each cell, the values represent the number of unique entities, for example the cells in the Assocs row represent the number of unique associations (rsids). However, some values can be replicated across the figure because a GWAS study may have researched several of the disease areas. Additionally, there is some non-additivity between consecutive rows, namely Druggable Gene Priority (Drug Gene Priority) - Distance Rank (Dist Rank) and Drugs - Drug indication/Disease Phenotypes Comparison (Drug I/Disease P Comparison). In the case of the former, this is due to the same gene being further away from the associated variant in different studies, such that it falls into a different partition. For the latter, this is due to missing indications for some of the drugs, such that concordance or discordance could not be assigned. The estimated number of samples (Est. N) is the sum of all the cases involved in the respective studies.