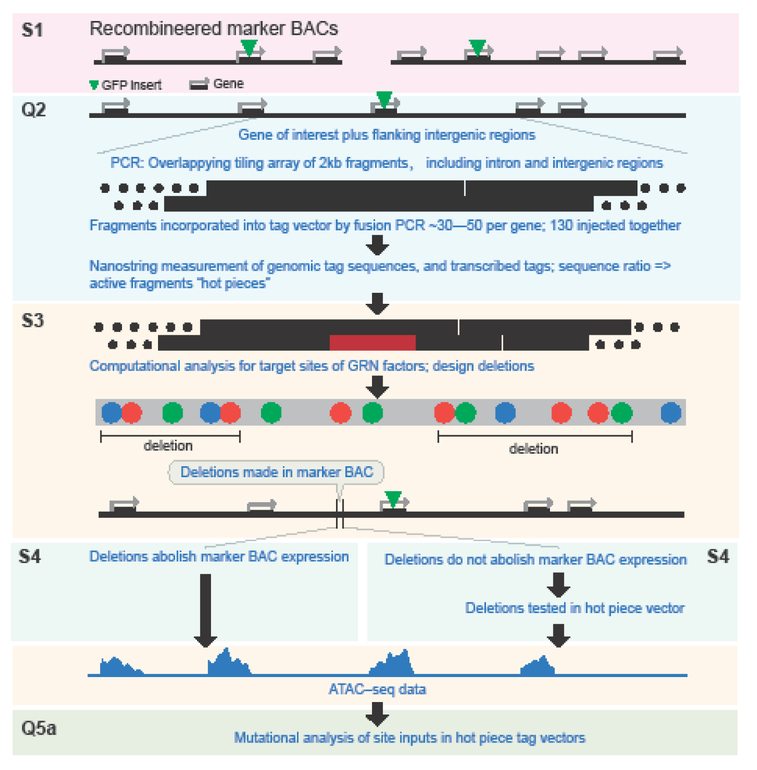
Fig.1.
Sequence of analytical procedures. Four or five successive sets of functional observation were made on the regulatory system of each gene. This was done either by microscopic observation of spatial expression of the GFP marker in individual embryos, in which the fraction of embryos examined was also recorded (S); or by quantitation of expression per construct using the multiplexed tag system in co-injected batches of embryos, measured by Nanostring as described in text (Q). At the first step, S1, efficacy of recombineered marker BACs was established. At the second, Q2, the whole intronic and intergenic region of each gene was segmented, scanned for activity, and one or more 2kb fragments displaying transcriptional activity (“hot pieces”) were identified. At the third, S3, spatial expression of marker BACs bearing specific deletions made within the putative regulatory regions (hot piece, marked as red rectangle) were assessed for spatial expression and fraction of embryos expressing, compared to controls. In most cases localized deletions were found which abolished marker BAC activity, and the analysis of these genes proceeded to step Q5. At the fourth step, S4, in cases where individual deletions failed to abolish BAC expression, these deletions were tested for spatial expression in the context of the individual active 2kb fragments. In the final step Q5, site clusters revealed to be functional in steps S3 or S4 were subjected en masse to site specific mutational analysis.