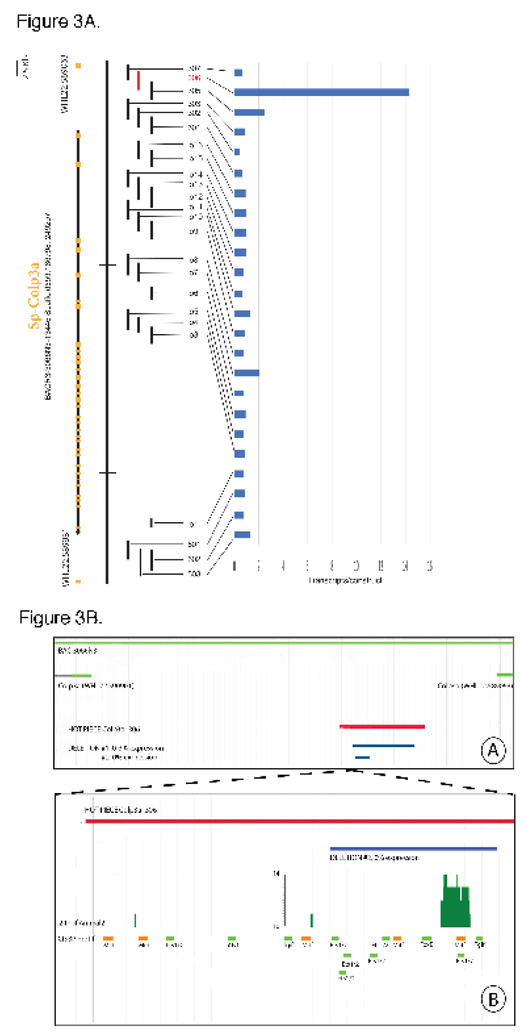
Fig.3.
Example of high throughput tag vector screen for cis-regulatory activity in a marker BAC. As described in text marker BACs, here containing the Colp3a gene, were segmented into ~2kb fragments forming an overlapping tiling array (black rectangles). Large exonic regions were excluded. Gene structure (exons) are represented by yellow boxes on the black (transcriptome) line. Flanking the black (transcriptome) line is the relevant region of the BAC, i.e., the gene plus the flanking upstream and downstream regions. The flanking yellow rectangles indicate the next gene upstream and downstream in the BAC. For identities of surrounding genes and BAC maps see http://www.echinobase.org/Echinobase/. Each 2kb fragment shown as a vertical bar was incorporated in a tag vector. The mixture of all 2kb-Tag vectors from 8 genes were injected into fertilized eggs. Transcribed tag DNA and genomically incorporated tag DNA for all vectors were measured simultaneously by Nanostring. The ratio of these values, which indicates the transcriptional activity driven by the sequence in each respective vector normalized to the amount of vector DNA incorporated, are shown at right in the histogram (Fig.3A). 2kb fragments that are thus revealed to include active cis-regulatory modules are shown in red.
Diagrams redrawn from JBrowse screen captures showing various features mapped to the reference genome sequence (Fig.3B). The BAC is shown in green. The red line depicts the DNA sequence of the active fragment “hot piece” in the BAC. Deleted regions are shown in blue below it. Symbols identifying the deletions are the ones used in the test and tables. Vertical bars represent the position and number of ATACseq reads mapped. The position of the modified CIS-BP transcription factor binding sites are indicated in light green. Mutated sites are indicated in orange.