Between 2003 and 2016, transmitted HIV-1 drug resistance has increased in a large California clinic population. However, the most commonly transmitted drug-resistance mutations reduce susceptibility primarily to drugs that are no longer frequently used.
Keywords: HIV-1 transmission, HIV-1 drug resistance, mutation, reverse transcriptase, protease
Abstract
Background
There are few large studies of transmitted drug resistance (TDR) prevalence and the drug resistance mutations (DRMs) responsible for TDR in the United States.
Methods
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) and protease sequences were obtained from 4253 antiretroviral therapy (ART)–naive individuals in a California clinic population from 2003 to 2016. Phylogenetic analyses were performed to study linkages between TDR strains and selection pressure on TDR-associated DRMs.
Results
From 2003 to 2016, there was a significant increase in overall (odds ratio [OR], 1.05 per year [95% confidence interval {CI}, 1.03–1.08]; P < .001) and nonnucleoside RT inhibitor (NNRTI)–associated TDR (OR, 1.11 per year [95% CI, 1.08–1.15]; P < .001). Between 2012 and 2016, TDR rates to any drug class ranged from 15.7% to 19.2%, and class-specific rates ranged from 10.0% to 12.8% for NNRTIs, 4.1% to 8.1% for nucleoside RT inhibitors (NRTIs), and 3.6% to 5.2% for protease inhibitors. The thymidine analogue mutations, M184V/I and the tenofovir-associated DRMs K65R and K70E/Q/G/N/T accounted for 82.9%, 7.3%, and 1.4% of NRTI-associated TDR, respectively. Thirty-seven percent of TDR strains clustered with other TDR strains sharing the same DRMs.
Conclusions
Although TDR has increased significantly in this large cohort, many TDR strains are unlikely to influence the activity of currently preferred first-line ART regimens. The high proportion of DRMs associated with infrequently used regimens combined with the clustering of TDR strains suggest that some TDR strains are being transmitted between ART-naive individuals.
In the United States, routine human immunodeficiency virus type 1 (HIV-1) genotypic resistance testing at the time of diagnosis or prior to starting antiretroviral therapy (ART) began about 15 years ago [1, 2]. Although there have been several US studies of transmitted drug resistance (TDR) prevalence, few large studies have characterized the evolution of TDR, the drug resistance mutations (DRMs) responsible for TDR, and the predicted clinical significance of these DRMs [3–11].
In this study, we examined the changing prevalence of TDR from 2003 to 2016 in a large clinic population in a region with high ART coverage. We also characterized the specific DRMs responsible for TDR, and the likely clinical significance of TDR in an era in which nonnucleoside reverse transcriptase inhibitor (NNRTI)–containing regimens are no longer preferred regimens for initial therapy, and in which thymidine analogues are rarely used. We performed phylogenetic analyses to determine how much TDR emanates from established circulating drug-resistant strains as opposed to multiple independent episodes of acquired drug resistance and to identify selection pressures at DRM positions in viruses from ART-naive individuals.
METHODS
Study Cohort and Virus Samples
The study cohort comprised all ART-naive adults in Kaiser Permanente Northern California (KPNC) undergoing dideoxynucleotide genotypic resistance testing of HIV-1 reverse transcriptase (RT) and protease between January 2003 and December 2016 at the Stanford University Healthcare Diagnostic Virology Laboratory. KPNC is estimated provide care to approximately 25% of the insured population in Northern California [12]. Cohort individuals were characterized by age, gender, race, HIV acquisition risk factor, and baseline plasma HIV-1 RNA level and CD4 count. For ART-naive individuals having >1 resistance test, the virus sequence of the first test was analyzed. Sequences from 13 individuals not encompassing protease positions 10–90 and RT positions 40–240 were excluded. The Stanford University and KPNC institutional review boards approved this study.
Prevalence and Temporal Trend of TDR
TDR was defined as the presence of 1 or more mutations from the World Health Organization 2009 list of 93 surveillance DRMs (SDRMs) at 43 positions, including 34 nucleoside reverse transcriptase inhibitor (NRTI)–, 19 NNRTI-, and 40 protease inhibitor (PI)–associated DRMs [13]. We determined the proportion of individuals with TDR to each class and with multiclass TDR. We used generalized binomial logistic regression models to assess the relationship between sample year and TDR and calculated the odds ratio for yearly increases in TDR prevalence.
A subset of the NRTI-associated SDRMs were classified as thymidine analogue mutations (TAMs) including M41L, D67N/G, K70R, L210W, T215Y/F, K219Q/E/R/N, and the T215 revertants T215C/D/E/I/S/V (which evolve from T215F/Y in the absence of selective drug pressure). Several additional DRMs not on the SDRM list were analyzed including (1) the primarily tenofovir disoproxil fumarate (TDF)–selected DRMs A62V, K65N, and K70G/N/Q/S/T [14] and (2) the primarily rilpivirine (RPV)–selected DRMs E138A/G/K/Q, of which E138A is polymorphic, occurring in 1%–4% of viruses from ART-naive individuals [15, 16].
The Stanford HIV Drug Resistance Database (HIVDB) genotypic resistance interpretation program was used to quantify the clinical impact of DRMs on the NRTIs abacavir (ABC), TDF, zidovudine (ZDV), and the cytosine analogues lamivudine and emtricitabine (3TC/FTC); the NNRTIs efavirenz (EFV), RPV, and etravirine (ETR); and the pharmacologically boosted PIs lopinavir/ritonavir (LPV/r), atazanavir (bATV), and darunavir (bDRV) [17].
Phylogenetic Analyses
We performed 2 phylogenetic analyses to characterize TDR transmission dynamics. The first, which included solely ART-naive individuals, examined the extent of sequence similarity among TDR strains. The second, which included both ART-naive and -experienced individuals, determined the proportion of TDR strains that were similar to strains from ART-experienced individuals. For both phylogenetic analyses we concatenated the 297 protease nucleotides and the 900 nucleotides encompassing RT positions 1–300, masked ambiguous nucleotides and those encoding SDRMs, and used IQTREE to create a maximum likelihood tree using a generalized time-reversible substitution model with site-to-site rate variation modeled by the discrete gamma distribution with 4 rate classes and an invariant proportion (GTR + I + G4), and generated bootstrap support using 1000 replicates [18].
For the first analysis, we used the complete set of sequences from the ART-naive cohort. Following tree construction, we used ClusterPicker to identify sequence clusters defined as sequences of subtrees having a maximum pairwise distance between sequences <0.02 and a bootstrap value >70% [19]. We then determined the proportion of TDR viruses in a cluster containing other viruses sharing the same SDRMs.
For the second analysis, we combined the sequences from the ART-naive cohort with sequences from ART-experienced individuals in KPNC who underwent resistance testing at Stanford University Healthcare since such testing began in 1998. For this analysis, we used a higher pairwise distance threshold of 0.04, albeit with a stricter bootstrap value of ≥90. The higher distance threshold and stricter bootstrap value defined clades in which all leaves shared a common ancestor but, as a result of the higher distance threshold, were less likely to be closely related epidemiologically than individuals in the ART-naive clusters. We then determined the proportion of individuals with TDR that had viruses in the same clade as a virus sequence, with the same SDRMs, from an ART-experienced individual with acquired drug resistance.
For the largest TDR cluster, we performed Bayesian phylogenetic inference of time-measured trees using Markov chain Monte Carlo (MCMC) sampling implemented in BEAST (Bayesian Evolutionary Analysis Sampling Trees) version 1.8.4 software [20]. The substitution process was modeled according to the GTR + I + G4 model and an exponential growth model was specified as coalescent tree prior. Independent MCMC analyses were run for 10 million generations, sampling every 5000 generations, and the first 10% of the samples was discarded as burn-in before combining the samples. The runs were investigated based on effective sample size calculated using Tracer [21]. Then a maximum clade credibility tree was selected from the posterior tree distribution and visualized using FigTree [22].
To determine if viral lineages in ART-naive individuals underwent diversifying positive selection at SDRM sites, we applied the fixed effects likelihood test in HyPhy version 2.3.10. The test was restricted only to terminal branches leading to sequences from ART-naive individuals, or those internal branches whose descendants were all ART-naive, in the phylogenetic tree containing sequences from both ART-naive and -experienced individuals [23].
RESULTS
Study Cohort
Between January 2003 and December 2016, 4253 HIV-1–infected ART-naive individuals underwent genotypic resistance testing. The median number of ART-naive individuals per year was 304 (range, 178–355). The median age was 39.5 and 90.5% were male (Table 1). The cohort composition was 42.5% white, 20.9% African American, 19.2% Latino, 8.0% Asian, 0.4% American Indian/Alaska Native, and 9.0% unknown. The HIV-1 acquisition risk factors included men who have sex with men (59.2%), heterosexual contact (18.1%), bisexual contact (10.4%), intravenous drug use (6.2%), receipt of transfusion (0.4%), and perinatal infection (0.1%). Acquisition risk factors were unrecorded for 5.6% of the population.
Table 1.
Association of Study Cohort Characteristics With Transmitted Drug Resistance
| Characteristic | Overall (N = 4253) | Wild-type (n = 3662) | TDR (n = 591) | P Valuea |
|---|---|---|---|---|
| Age, y, mean | 39.5 | 39.6 | 39.2 | .4 |
| HIV-1 RNA, mean, log copies/mL | 4.5 | 4.5 | 4.4 | 1 |
| CD4 count, mean, cells/µL | 376 | 372 | 402 | .02 |
| Male, % | 90.5 | 90.5 | 90.3 | 1 |
| Race/ethnicity, % | ||||
| White | 42.5 | 43.0 | 39.3 | .1 |
| Black/African American | 20.9 | 21.5 | 17.0 | .01 |
| Hispanic/Latino | 19.2 | 18.9 | 21.4 | .2 |
| Asian | 8.0 | 7.5 | 11.5 | .001 |
| American Indian/Alaska Native | 0.4 | 0.4 | 0.5 | 1 |
| Unknown | 9.0 | 8.7 | 10.3 | .2 |
| Transmission risk, % | ||||
| MSM | 59.2 | 59.2 | 59.2 | 1 |
| Heterosexual contact | 18.1 | 18.1 | 18.3 | .95 |
| Bisexual contact | 10.4 | 10.4 | 10.7 | .91 |
| Intravenous drug useb | 6.2 | 6.2 | 5.8 | .73 |
| Transfusion | 0.4 | 0.5 | 0 | .17 |
| Perinatal | 0.1 | 0.1 | 0.3 | .17 |
| Unknown | 5.6 | 5.5 | 5.8 | .89 |
| Subtype, % | ||||
| Subtype B | 95.3 | 94.9 | 97.6 | .005 |
| Subtype C | 1.3 | 1.3 | 1.0 | .7 |
| CRF01_AE | 1.2 | 1.4 | 0.2 | .02 |
| CRF02_AG | 0.5 | 0.6 | 0.3 | .7 |
| Subtype A | 0.4 | 0.5 | 0 | .2 |
| Other subtypes/CRFs | 1.3 | 1.3 | 0.9 | .4 |
Except for continuous data, given as means, data are shown as percentages of the total number of individuals indicated in the column headers.
Abbreviations: CRF, circulating recombinant form; HIV-1, human immunodeficiency virus type 1; MSM, men who have sex with men; TDR, transmitted drug resistance.
aStudent t test was used for comparisons of continuous data (age, HIV-1 RNA, and CD4 cell count). The χ2 test was used for comparisons of categorical data.
bIntravenous drug users included individuals with or without MSM as a risk factor.
At the time of genotypic resistance testing, the median CD4 count was 349 cells/µL (interquartile range [IQR], 180–527 cells/µL) and the median plasma HIV-1 RNA level was 4.5 log copies/mL (IQR, 4.0–5.1 log copies/mL). One hundred seventy-nine (4.2%) individuals had repeated genotypic resistance tests before starting ART. Subtype B accounted for 95.3% of viruses. The remaining 4.7% included subtype C (1.3%), circulating recombinant form (CRF) 01_AE (1.2%), CRF02_AG (0.5%), A (0.4%), and other subtypes and CRFs (1.3%).
The 4253 sequences contained a median of 0.67% mixtures per nucleotide (IQR, 0.17%–1.33%). There was a yearly 1.06-fold (95% confidence interval [CI], 1.05–1.08; P < .001) increase in the odds of having a sequence with <0.5% mixtures, a proxy for recent infection [24–26], with 29.8% of 2003 and 52.3% of 2016 samples having <0.5% mixtures. Consistent with the concept that over time the cohort underwent resistance testing earlier during infection, there was also a significant yearly increase in CD4 cell count (regression coefficient of 10.9 cells/µL; P < .001). The mean CD4 count in 2003 was 259 cells/µL and in 2016 was 419 cells/µL.
TDR Prevalence and Temporal Trends
Over the 14 year-study period, 591 of 4253 (13.9%) individuals had TDR. The overall proportions with NNRTI, NRTI, PI, and multiclass TDR were 7.2%, 5.8%, 3.2%, and 1.9%, respectively. Table 1 shows that subtype B and Asian race were associated with an increased TDR risk whereas African American race was associated with a reduced TDR risk. TDR risk was not significantly influenced by age, gender, acquisition risk factor, CD4 count, or plasma HIV-1 RNA level.
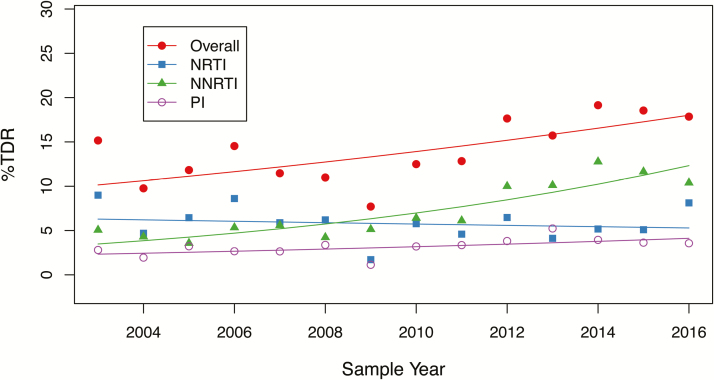
Figure 1 shows that there was a significant yearly increase in overall (odds ratio [OR], 1.05 [95% CI, 1.03–1.08]; P < .001) and NNRTI-associated TDR (OR, 1.11 [95% CI, 1.08–1.15]; P < .001). There was also weakly significant increases in PI-associated (OR, 1.05 [95% CI, 1.0–1.09]; P = .05) and multiclass TDR (OR, 1.09 [95% CI, 1.03–1.16]; P = .005). There was no significant temporal change in NRTI-associated TDR (OR, 0.99 [95% CI, .95–1.02]; P = .4). Supplementary Table 1 lists the proportions of individuals with TDR by drug class for each study year.
Figure 1.
Temporal trends in the yearly proportion of individuals with transmitted drug resistance, by drug class. The fitted lines show the effect of sample years in generalized binomial logistic regression models. Abbreviations: NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; TDR, transmitted drug resistance.
SDRMs Responsible for TDR
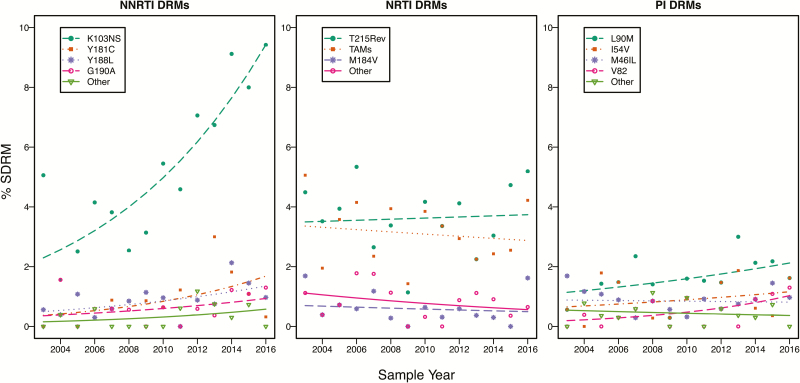
Among viruses from the 591 individuals with TDR, there were a total of 960 SDRMs (1.62 per individual): 387 (65.5%) had 1 SDRM, 128 (21.7%) had 2 SDRMs, and 76 (12.8%) had ≥3 SDRMs. Figure 2 displays the temporal trends of the most common NNRTI, NRTI, and PI SDRMs. Supplementary Tables 2–4 contain the complete list of SDRMs and their proportions by ART class.
Figure 2.
Temporal trends in the yearly proportion of individuals with the most commonly detected nucleoside reverse transcriptase inhibitor, nonnucleoside reverse transcriptase inhibitor, and protease inhibitor surveillance drug resistance mutations. The fitted lines show the effect of sample years in generalized binomial logistic regression models. Abbreviations: DRM, drug resistance mutation; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; SDRM, surveillance drug resistance mutation; TAM, thymidine analogue mutation.
The mutations K103N/S, Y181C, Y188L, and G190A accounted for 88.5% of the 348 NNRTI SDRMs. The mutations E138A/G/K/Q, which are associated with reduced RPV susceptibility but are not on the SDRM list, occurred in 99 individuals, including 90 without an SDRM. The polymorphic mutation E138A accounted for 87 of these 99 mutations [15, 16]. For the entire population, the proportion with predicted low-, intermediate-, or high-level NNRTI resistance was 7.8% for EFV, 5.1% for RPV, and 1.6% for ETR.
Of the 409 NRTI SDRMs, 82.9% were TAMs (including T215 revertant mutations). However, the primary TAMs T215F/Y were present in just 5 individuals. M184V/I occurred in 20 individuals, comprising 7.3% of NRTI SDRMs. The non-TAMs K65R, L74V/I, Y115F, and Q151M each occurred in just 1 or 2 individuals. The TDF-associated SDRM K70E did not occur. K70N/T/S, which are not on the SDRM but are associated with low-level TDF resistance, occurred in 3 individuals including 1 without an SDRM. A62V, a non-SDRM TDF-associated accessory DRM, occurred in 10 individuals. For the entire population, the proportion with predicted low-, intermediate-, or high-level NRTI resistance was 4.9% for ZDV, 2.8% for ABC, 2.2% for TDF, and 0.7% for 3TC/FTC.
Of the 203 PI-associated SDRMs, the most common were L90M (33.5%), M46I/L (19.7%), I54V (10.8%), V82A/L/T (10.4%), and D30N plus N88D (9.8%). The next most common SDRMs, V32I, I50V/L, L76V, and I84V, each occurred in ≤5 individuals. For the entire population, the proportion with predicted low-, intermediate-, or high-level PI resistance was 2.2% for bATV, 2.1% for LPV/r, and 0.3% for bDRV.
Virus Sequence Clustering
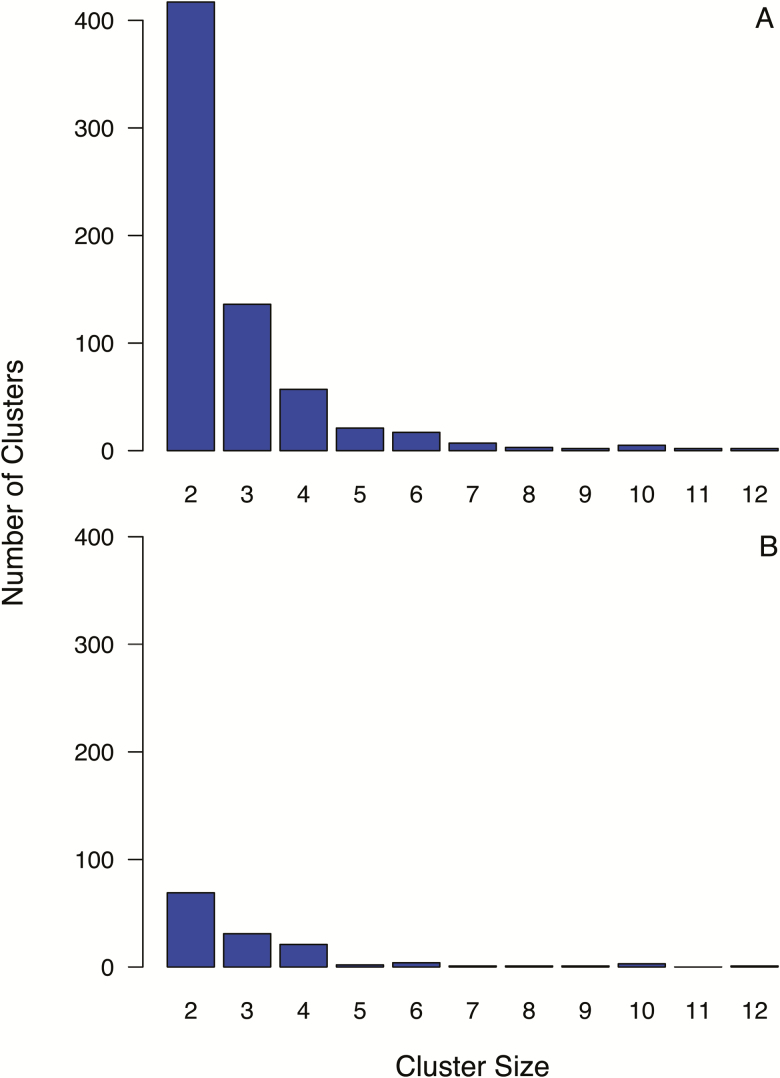
The median uncorrected intrasubtype pairwise distance between sequences in the ART-naive cohort was 0.055 (IQR, 0.047–0.062). Of these sequences, 43.8% (1864) were present in 1 of 669 clusters. Of these, 134 contained 1 or more sequences with an SDRM, comprising 47.0% of the 591 individuals with TDR. Figure 3A and 3B show the distribution of the number of clusters by cluster size for all 669 clusters and for the 134 clusters containing 1 or more sequences with an SDRM.
Figure 3.
Number of clusters by cluster size for the complete antiretroviral therapy–naive dataset of 4235 virus sequences (A) and for the clusters containing 1 or more of the 591 viruses with transmitted drug resistance (B). Sequence clusters were defined as sequences (leaves) of subtrees having a maximum pairwise uncorrected distance between leaves of ≤0.02 and a bootstrap value of ≥70%.
Of the 669 clusters, 97.3% comprised subtype B viruses; 2.7% (n = 18) comprised viruses belonging to other subtypes including C (6), CRF01_AE (6), CRF02_AG (2), A (1), D (1), F (1), and G (1). One CRF01_AE cluster contained 12 viruses without SDRMs. The remaining non–subtype B clusters contained 2–3 viruses including 2 clusters with SDRMs.
Composition of Clusters Containing TDR Viruses
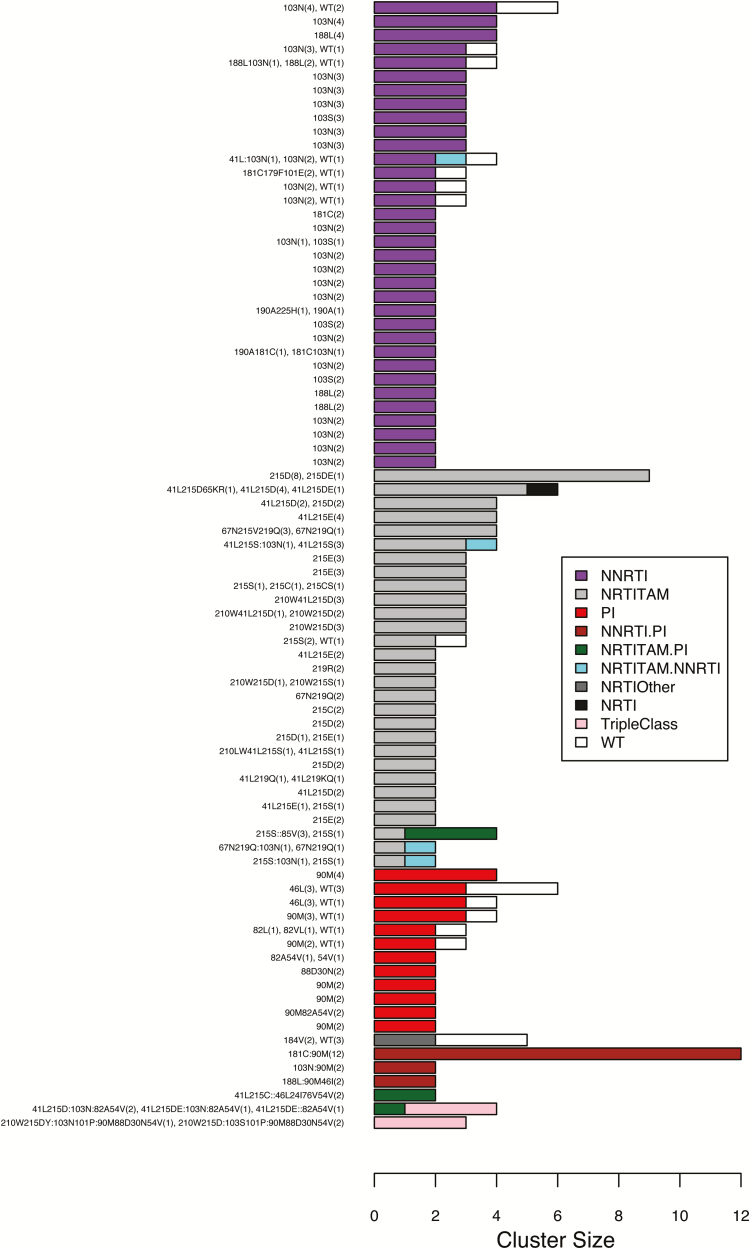
Of the 134 clusters containing viruses with an SDRM, 82 contained ≥2 viruses sharing 1 or more SDRMs (Figure 4). Within these 82 clusters, the median cluster size was 2 (range, 2–12) and the median pairwise distance was 0.0064. The median range in years from the earliest to most recent sample was 4.0 years (IQR, 2.0–6.8 years). In 23.2% (n = 19) of the 82 TDR clusters, each virus was obtained within the same 12-month period.
Figure 4.
Composition of surveillance drug resistance mutation (SDRM) patterns of the 82 clusters containing ≥2 viruses sharing ≥1 SDRM. Clusters were defined as including viruses with a maximum pairwise distance ≤0.02 and bootstrap support value ≥70%. Overall, 68 (82.9%) of the clusters consisted entirely of viruses with SDRMs and 55 (67.1%) were homogeneous (ie, containing the same SDRMs). Abbreviations: NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NRTITAM, nucleoside reverse transcriptase inhibitor thymidine analogue mutation; PI, protease inhibitor; WT, wild-type.
Overall, 68 of 82 (82.9%) clusters in Figure 4 consisted entirely of viruses with SDRMs including 27 containing 64 NNRTI TDR viruses, 24 containing 71 NRTI TDR viruses, 7 containing 16 PI TDR viruses, and 10 containing 37 viruses with SDRMs conferring resistance to >1 drug class. The remaining 14 clusters contained 1 or more viruses without SDRMs in addition to viruses with an SDRM. Overall, 55 of the 82 (67.1%) clusters were homogeneous in that they contained identical SDRMs; 27 (32.9%) were heterogeneous containing 1 or more viruses without SDRMs or with different, albeit overlapping, SDRMs.
TDR Viruses Similar to Those From ART-experienced Individuals
The phylogenetic analysis of the sequences from the 4253 ART-naive individuals combined with 4064 sequences from 3155 ART-experienced individuals showed that 83 viruses from the 591 (14%) ART-naive individuals with TDR were in the same clade as a virus from an ART-experienced individual whose virus contained each of the SDRMs as the TDR virus. The median uncorrected genetic distance between these 83 virus pairs was 0.009 (IQR, 0.003–0.015).
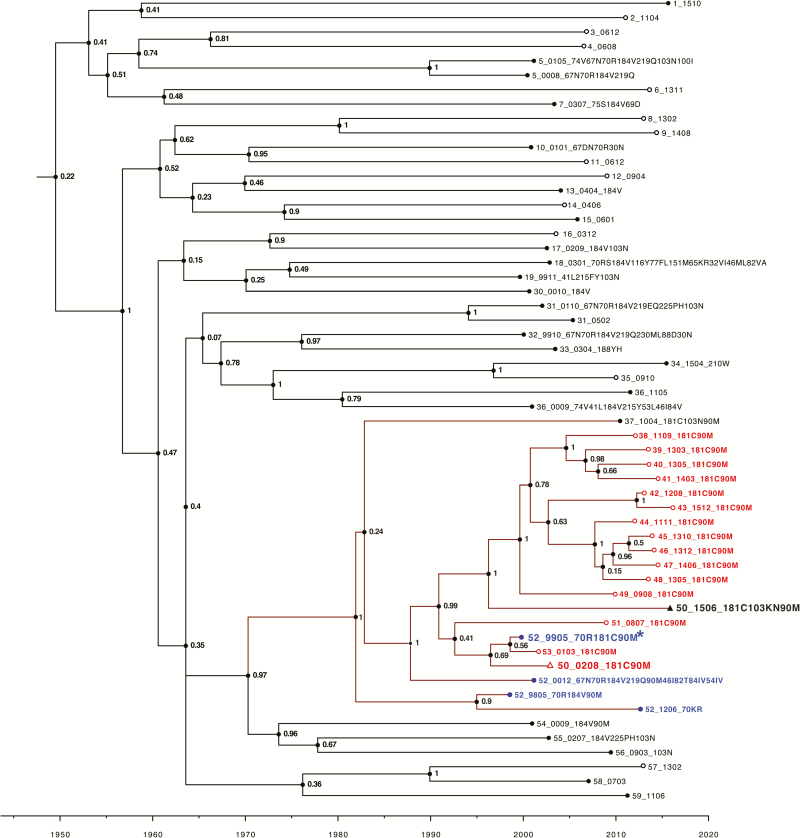
Figure 5 contains a time-scaled Bayesian phylogenetic tree that illustrates in red the likely origin of the cluster of 12 viruses containing the NNRTI DRM Y181C plus the PI DRM L90M. The drug-resistant virus first emerged in 1999 in an individual who received sequential therapy with several drugs including ZDV, 3TC, stavudine, nevirapine, and saquinavir. This individual had 4 genotypes performed between 1998 and 2012, each of which is indicated in blue.
Figure 5.
Time-scale analysis of the large virus cluster containing viruses with the nonnucleoside reverse transcriptase inhibitor resistance mutation Y181C and the protease inhibitor resistance mutation L90M. Viruses were labeled with a randomly generated person identifier (PID), the sample year and month in four digits (two-digit sample year and two-digit month), and the viruses’ list of surveillance drug resistance mutations delimited by “_”. The values at the nodes represent posterior support values for the clusters obtained using Markov chain Monte Carlo sampling implemented in BEAST version 1.8.4 software. Leaf nodes for sequences from antiretroviral therapy (ART)–naive individuals are unfilled circles whereas those from ART-experienced individuals are filled circles. Viruses from 16 ART-naive individuals with Y181C + L90M are indicated in red, and 4 viruses with the same mutations from an ART-experienced individual are indicated in blue (PID 52). This ART-experienced individual appeared to acquire Y181C + L90M plus K70R in 1999 (sequence followed by an asterisk). One individual (PID 50) was also primarily infected with this virus as a 2002 sequence obtained prior to therapy contained Y181C + L90M (indicated in red with an open triangle) and later developed K103N (indicated in black with a closed triangle) after receiving multiple nucleoside reverse transcriptase inhibitor regimens followed by tenofovir/emtricitabine/efavirenz in combination with atazanavir/ritonavir and then raltegravir. There were an additional 3 individuals whose viruses likely originated from this virus strain but were not in the same cluster because their sequence differed from the 12 clustered viruses by >2.0%.
Positional Selection Pressure in Protease and RT
The fixed-effects likelihood test for selection pressure on the branches leading to the sequences from ART-naive individuals identified 29 codons in protease and RT that were subject to diversifying positive selection (P ≤ .01), but there was no evidence of positive selection at any of the 43 SDRM positions. Each of these 43 codons was inferred to be subject to purifying selection, and at 40 of the codons this finding was statistically significant (P ≤ .01, likelihood ratio test). Supplementary Table 5 summarizes the fixed-effects likelihood test results for all 43 SDRM positions.
DISCUSSION
In the 14 years since pretreatment HIV genotypic resistance testing became the standard of care in this Northern California clinic-based population, there has been a progressive increase in TDR. The increase occurred in multiple demographic groups and in individuals with diverse HIV-1 acquisition risk factors. However, because most TDR strains had NNRTI-associated DRMs or NRTI-associated TAMs, they are unlikely to influence the activity of the preferred first-line ART regimens, which infrequently include NNRTIs and very rarely include thymidine analogues. Our finding of an ongoing TDR increase is consistent with the high TDR levels observed in other North American studies [4–6, 8, 27]. The finding that NNRTI-associated TDR has been increasing is consistent with the global TDR trends [28, 29].
Consistent with a previous meta-analysis, a small subset of NNRTI-associated SDRMs accounted for the vast majority of NNRTI TDR [29]. Although the spectrum of NRTI-associated SDRMs was diverse, >80% of NRTI TDR cases were caused by TAMs, of which those other than T215Y/F may have little impact on currently used NRTIs [30]. The rarity of K65R, other less common TDF-associated mutations, and the primary TAMs T215Y/F indicates that transmitted TDF resistance is unusual. Moreover, the evolution of US treatment guidelines toward first-line regimens that include an integrase strand transfer inhibitor (INSTI) or bDRV means that the preferred first-line regimens are highly active in most patients with TDR [31].
The prevalence of TDR in chronically infected individuals is generally lower than in acutely infected individuals because in the years following initial infection, the least fit DRMs revert to wild type [32–35]. Indeed, our selection analyses shows that within the combined phylogeny of sequences from ART-naive and ART-experienced individuals, natural selection maintained existing amino acids at the SDRM sites along the ART-naive branches. Thus, despite their ability to persist in ART-naive populations, the overall trend for many SDRMs is toward a gradual reversion to wild type. Some of the increase in TDR prevalence may also have resulted from the trend to perform genotypic resistance testing earlier in infection as evidenced by the cohort’s progressively lower proportion of sequence mixtures and higher CD4 counts.
Endemic TDR strains emanating from a single instance of ART-selection pressure that spread among ART-naive individuals have different implications from TDR strains emanating from independent episodes of ART-selection pressure in different ART-experienced individuals. Endemic strains may carry a greater risk of ongoing transmission consistent with their ability to persist in a population in the absence of selective drug pressure [29]. However, they may also be less likely to harbor additional DRMs as the process of ongoing transmission among ART-naive individuals would be expected to filter out minority variants that may have originally been transmitted from an ART-experienced individual [36, 37].
The high proportion of DRMs associated with infrequently used ART regimens combined with the clustering of many TDR strains suggests that some proportion of TDR strains represent established drug-resistant lineages transmitted among ART-naive individuals. This hypothesis is consistent with findings from other countries with mature HIV-1 epidemics such as Switzerland and the United Kingdom, where it has been estimated that most TDR cases are transmitted from ART-naive individuals [34, 36, 38, 39]. Our dataset, however, is limited by the fact that KPNC provides care to about 25% of the insured population in Northern California [12], making it likely that many transmission events occurred with individuals not within our cohort and not captured in our phylogenetic analyses.
In conclusion, this is one of the largest studies of the trends and mutation patterns associated with HIV-1 TDR in the United States. The finding that a large proportion of TDR strains contain DRMs associated with regimens now used infrequently is consistent with the concept that many TDR cases are transmitted between ART-naive individuals. Although the frequency of transmitted INSTI resistance has been extremely rare in the United States [40], ongoing surveillance is required as INSTIs have been increasingly used for both first-line and salvage ART.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. The complete set of sequence data from 4253 antiretroviral therapy (ART-naive) individuals has been submitted to GenBank. Of the 4253 ART-naive individuals, sequences from 1144 individuals had been previously deposited and are available in GenBank and their accession numbers are listed in the Supplementary Materials. The sequences from the remaining individuals have been submitted to GenBank (MG941022–MG944215).
Financial support. R. W. S. and S. Y. R. were supported in part by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) (award number AI068581). D. S. C. was supported by research funding from the Bristol-Myers Squibb Virology Fellows Research Program and by the KL2 Mentored Career Development Award of the Stanford Clinical and Translational Science Award to Spectrum (award numbers NIH KL2 TR 001083 and NIH UL1 TR 001085). J. L. M. was supported in part by the NIAID (award number K01 122853). S. L. K. P. was supported in part by grants R01 GM093939 (NIH/National Institute of General Medical Sciences [NIGMS]), R01 AI134384 (NIH/NIAID), and U01 GM110749 (NIH/NIGMS).
Potential conflicts of interest. R. W. S. has received grants from Gilead Sciences, Janssen Pharmaceuticals, and Vela Diagnostics, and personal fees from ViiV and Abbott Diagnostics. D. S. C. has received research funding from Bristol-Myers Squibb. J. L. M. has received research grant support from Merck. M. S. has received grants from Pfizer, Merck, and Gilead. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Sax PE, Islam R, Walensky RP, et al. . Should resistance testing be performed for treatment-naive HIV-infected patients? A cost-effectiveness analysis. Clin Infect Dis 2005; 41:1316–23. [DOI] [PubMed] [Google Scholar]
- 2. Hirsch MS, Brun-Vézinet F, Clotet B, et al. . Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA panel. Clin Infect Dis 2003; 37:113–28. [DOI] [PubMed] [Google Scholar]
- 3. Wheeler WH, Ziebell RA, Zabina H, et al. . Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. AIDS 2010; 24:1203–12. [DOI] [PubMed] [Google Scholar]
- 4. Banaez Ocfemia MC, Saduvala N, Oster AM, et al. . Transmitted HIV-1 drug resistance among men who have sex with men—11 U.S. jurisdictions, 2008–2011 [abstract 579]. In: 21st Conference on Retroviruses and Opportunistic Infections, Boston, MA, 3–6 March 2014. [Google Scholar]
- 5. Panichsillapakit T, Smith DM, Wertheim JO, Richman DD, Little SJ, Mehta SR. Prevalence of transmitted HIV drug resistance among recently infected persons in San Diego, CA 1996–2013. J Acquir Immune Defic Syndr 2016; 71:228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kassaye SG, Grossman Z, Balamane M, et al. . Transmitted HIV drug resistance is high and longstanding in metropolitan Washington, DC. Clin Infect Dis 2016; 63:836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gagliardo C, Brozovich A, Birnbaum J, et al. . A multicenter study of initiation of antiretroviral therapy and transmitted drug resistance in antiretroviral-naive adolescents and young adults with HIV in New York City. Clin Infect Dis 2014; 58:865–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aldous AM, Castel AD, Parenti DM; DC Cohort Executive Committee Prevalence and trends in transmitted and acquired antiretroviral drug resistance, Washington, DC, 1999–2014. BMC Res Notes 2017; 10:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taniguchi T, Nurutdinova D, Grubb JR, et al. . Transmitted drug-resistant HIV type 1 remains prevalent and impacts virologic outcomes despite genotype-guided antiretroviral therapy. AIDS Res Hum Retroviruses 2012; 28:259–64. [DOI] [PubMed] [Google Scholar]
- 10. Poon AF, Aldous JL, Mathews WC, et al. . Transmitted drug resistance in the CFAR network of integrated clinical systems cohort: prevalence and effects on pre-therapy CD4 and viral load. PLoS One 2011; 6:e21189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Youmans E, Tripathi A, Albrecht H, Gibson JJ, Duffus WA. Transmitted antiretroviral drug resistance in individuals with newly diagnosed HIV infection: South Carolina 2005–2009. South Med J 2011; 104:95–101. [DOI] [PubMed] [Google Scholar]
- 12. Gordan N. Similarity of the adult Kaiser Permanente membership in Northern California to the insured and general population in Northern California: statistics from the 2011 California Health Interview Survey. 2015. Available at: https://divisionofresearch.kaiserpermanente.org/projects/memberhealthsurvey/SiteCollectionDocuments/chis_non_kp_2011.pdf. Accessed 10 April 2018. [Google Scholar]
- 13. Bennett DE, Camacho RJ, Otelea D, et al. . Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009; 4:e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rhee SY, Varghese V, Holmes SP, et al. . Mutational correlates of virological failure in individuals receiving a WHO-recommended tenofovir-containing first-line regimen: an international collaboration. EBioMedicine 2017; 18:225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sluis-Cremer N, Jordan MR, Huber K, et al. . E138A in HIV-1 reverse transcriptase is more common in subtype C than B: implications for rilpivirine use in resource-limited settings. Antiviral Res 2014; 107:31–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rimsky L, Van Eygen V, Hoogstoel A, et al. . 96-week resistance analyses of rilpivirine in treatment-naive, HIV-1-infected adults from the ECHO and THRIVE phase III trials. Antivir Ther 2013; 18:967–77. [DOI] [PubMed] [Google Scholar]
- 17. Paredes R, Tzou PL, van Zyl G, et al. . Collaborative update of a rule-based expert system for HIV-1 genotypic resistance test interpretation. PLoS One 2017; 12:e0181357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ragonnet-Cronin M, Hodcroft E, Hué S, et al. . UK HIV Drug Resistance Database Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013; 14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007; 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rambaut A, Drummond AJ.. Tracer: MCMC trace analysis tool. 1.5 e. 2009. Available at: http://treebioedacuk/software/tracer. Accessed 1 June 2017. [Google Scholar]
- 22. Rambaut A. FigTree. 2017. Available at: http://treebioedacuk/software/figtree/. Accessed 1 June 2017. [Google Scholar]
- 23. Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 2005; 22:1208–22. [DOI] [PubMed] [Google Scholar]
- 24. Andersson E, Shao W, Bontell I, et al. . Evaluation of sequence ambiguities of the HIV-1 pol gene as a method to identify recent HIV-1 infection in transmitted drug resistance surveys. Infect Genet Evol 2013; 18:125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ragonnet-Cronin M, Aris-Brosou S, Joanisse I, et al. . Genetic diversity as a marker for timing infection in HIV-infected patients: evaluation of a 6-month window and comparison with BED. J Infect Dis 2012; 206:756–64. [DOI] [PubMed] [Google Scholar]
- 26. Kouyos RD, von Wyl V, Yerly S, et al. . Swiss HIV Cohort Study Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis 2011; 52:532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rocheleau G, Brumme CJ, Shoveller J, Lima VD, Harrigan PR. Longitudinal trends of HIV drug resistance in a large Canadian cohort, 1996–2016. Clin Microbiol Infect 2018; 24:185–91. [DOI] [PubMed] [Google Scholar]
- 28. Gupta RK, Gregson J, Parkin N, et al. . HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis 2018; 18:346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rhee SY, Blanco JL, Jordan MR, et al. . Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med 2015; 12:e1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Margot NA, Wong P, Kulkarni R, et al. . Commonly transmitted HIV-1 drug resistance mutations in reverse-transcriptase and protease in antiretroviral treatment-naive patients and response to regimens containing tenofovir disoproxil fumarate or tenofovir alafenamide. J Infect Dis 2017; 215:920–7. [DOI] [PubMed] [Google Scholar]
- 31. US Department of Health and Human Services Panel on Clinical Practices for Treatment of HIV Infection. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents (March 2018). Available at: http://aidsinfo.nih.gov/guidelines. Accessed 10 April 2018. [Google Scholar]
- 32. Jain V, Sucupira MC, Bacchetti P, et al. . Differential persistence of transmitted HIV-1 drug resistance mutation classes. J Infect Dis 2011; 203:1174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Castro H, Pillay D, Cane P, et al. . UK Collaborative Group on HIV Drug Resistance Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis 2013; 208:1459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang WL, Kouyos RD, Böni J, et al. . Swiss HIV Cohort Study Persistence of transmitted HIV-1 drug resistance mutations associated with fitness costs and viral genetic backgrounds. PLoS Pathog 2015; 11:e1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pingen M, Wensing AM, Fransen K, et al. . SPREAD Programme Persistence of frequently transmitted drug-resistant HIV-1 variants can be explained by high viral replication capacity. Retrovirology 2014; 11:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Drescher SM, von Wyl V, Yang WL, et al. . Swiss HIV Cohort Study Treatment-naive individuals are the major source of transmitted HIV-1 drug resistance in men who have sex with men in the Swiss HIV Cohort Study. Clin Infect Dis 2014; 58:285–94. [DOI] [PubMed] [Google Scholar]
- 37. Chaillon A, Nakazawa M, Wertheim JO, et al. . No substantial evidence for sexual transmission of minority HIV drug resistance mutations in men who have sex with men. J Virol 2017; 91. doi: 10.1128/JVI.00769-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mourad R, Chevennet F, Dunn DT, et al. . UK HIV Drug Resistance Database/Collaborative HIV, Anti-HIV Drug Resistance Network A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK. AIDS 2015; 29:1917–25. [DOI] [PubMed] [Google Scholar]
- 39. Ragonnet-Cronin ML, Shilaih M, Günthard HF, et al. . A direct comparison of two densely sampled HIV epidemics: the UK and Switzerland. Sci Rep 2016; 6:32251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hernandez AL, Ocfemia M, Saduvala N, et al. . HIV integrase genotypic testing and resistance in the United States—9 jurisdictions [abstract 978]. In: Conference on Retroviruses and Opportunistic Infections, Boston, MA, 13–16 February 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.