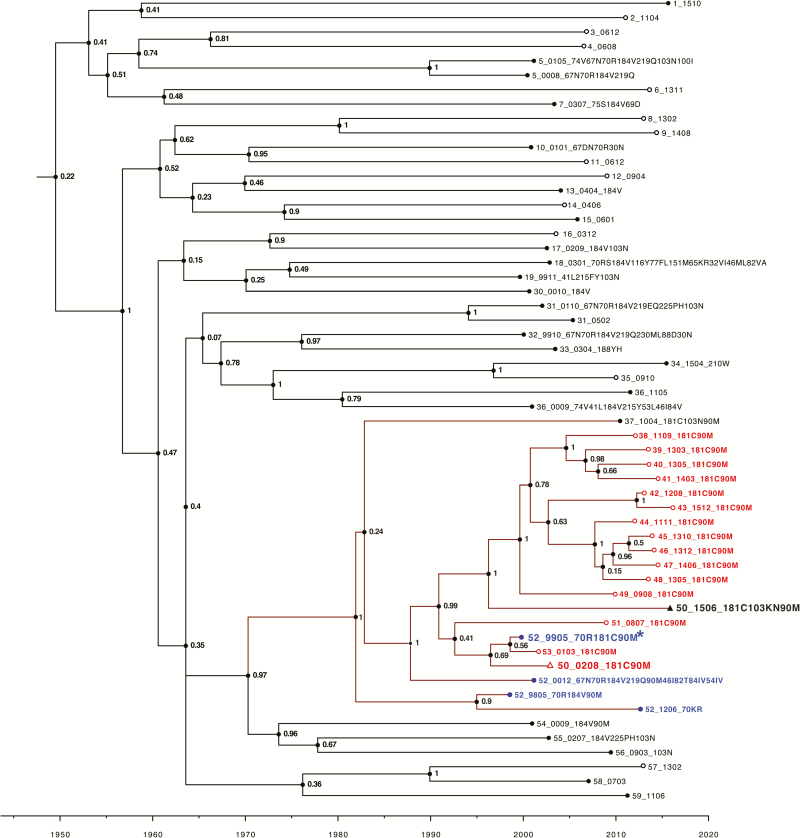
Figure 5.
Time-scale analysis of the large virus cluster containing viruses with the nonnucleoside reverse transcriptase inhibitor resistance mutation Y181C and the protease inhibitor resistance mutation L90M. Viruses were labeled with a randomly generated person identifier (PID), the sample year and month in four digits (two-digit sample year and two-digit month), and the viruses’ list of surveillance drug resistance mutations delimited by “_”. The values at the nodes represent posterior support values for the clusters obtained using Markov chain Monte Carlo sampling implemented in BEAST version 1.8.4 software. Leaf nodes for sequences from antiretroviral therapy (ART)–naive individuals are unfilled circles whereas those from ART-experienced individuals are filled circles. Viruses from 16 ART-naive individuals with Y181C + L90M are indicated in red, and 4 viruses with the same mutations from an ART-experienced individual are indicated in blue (PID 52). This ART-experienced individual appeared to acquire Y181C + L90M plus K70R in 1999 (sequence followed by an asterisk). One individual (PID 50) was also primarily infected with this virus as a 2002 sequence obtained prior to therapy contained Y181C + L90M (indicated in red with an open triangle) and later developed K103N (indicated in black with a closed triangle) after receiving multiple nucleoside reverse transcriptase inhibitor regimens followed by tenofovir/emtricitabine/efavirenz in combination with atazanavir/ritonavir and then raltegravir. There were an additional 3 individuals whose viruses likely originated from this virus strain but were not in the same cluster because their sequence differed from the 12 clustered viruses by >2.0%.