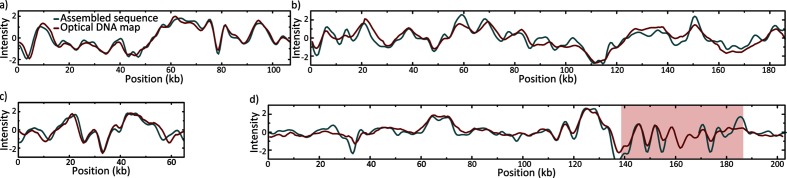
Fig. 1.
Comparison between the consensus of experimentally generated optical DNA maps (red) and theoretical optical DNA maps based on assembled sequences (teal) for novel plasmids (a) pEco70745_1 in E. coli CCUG 70745 (107 kb), (b) pKpn70747_1 in K. pneumoniae CCUG 70747 (186 kb), (c) pKpn70742_1 in K. pneumoniae CCUG 70742 (65 kb) and (d) pPmi70746_1 in Pr. mirabilis CCUG 70746 (experimental: 202 kb, theoretical: 192 kb). Intensity (y-axis) refers to the normalized emission intensity along the DNA molecule (x-axis), which correlates coarsely with the underlying DNA sequence. To aid the fitting of the barcodes in (d) – which are of different size – a gap has been introduced in the teal curve inside the red box. This box corresponds to a gene duplication and amplification region in pPmi70746_1 (see Fig. S1 for a histogram of individual plasmid lengths of pPmi70746_1).