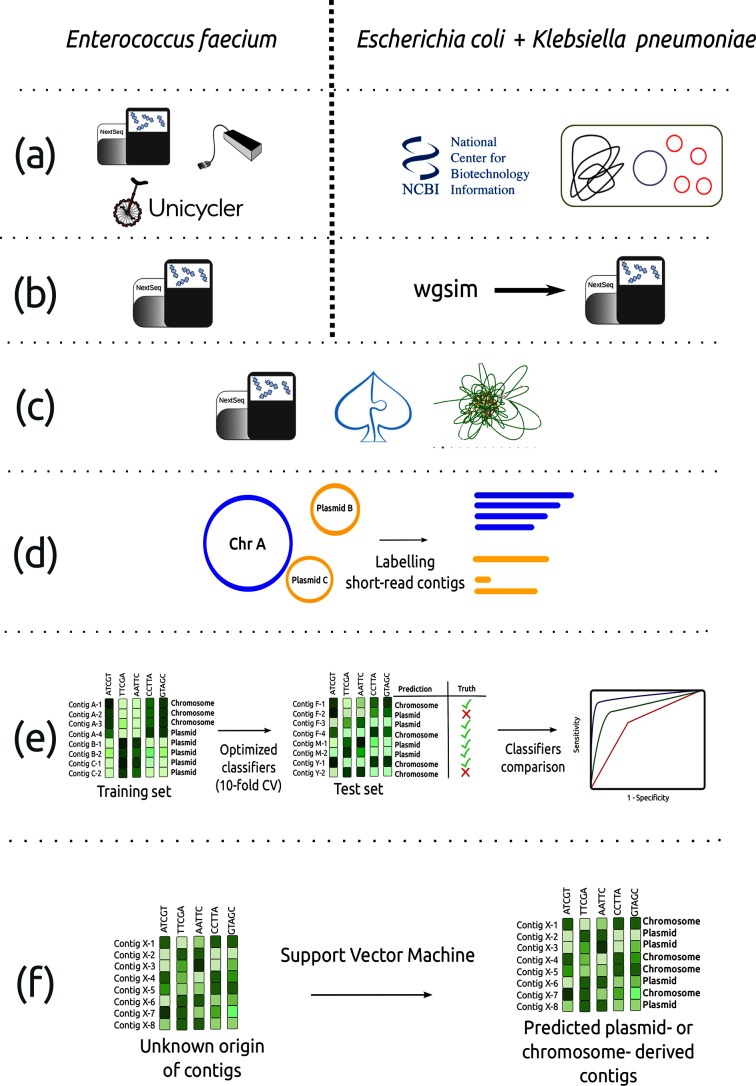
Fig. 1.
Workflow to create the plasmid models for Enterococcus faecium, Klebsiella pneumoniae and Escherichia coli. (a) For E. faecium, 62 Illumina-sequenced strains were selected for ONT sequencing and Unicycler was used to extend the number of complete genomes available for this species. For E. coli and K. pneumoniae, we downloaded complete genomes with plasmids associated from the Assembly Entrez NCBI database. (b) For E. coli and K. pneumoniae, we simulated reads with 50× coverage and no error rate using wgsim. (c) Illumina simulated and non-simulated reads were de novo assembled using SPAdes. (d) We mapped short-read contigs against complete genome sequences to define a reliable dataset of short-read contigs as plasmid or chromosome derived. (e) For each bacterial species, five machine-learning classifiers were trained (10-fold cross-validation) and compared using a specific bacterial species training and test set. (f) SVM models were implemented in mlplasmids and used to predict plasmid- and chromosome-derived sequences in isolates with only short-read WGS data available. The complete workflow is available from https://gitlab.com/sirarredondo/analysis_mlplasmids.