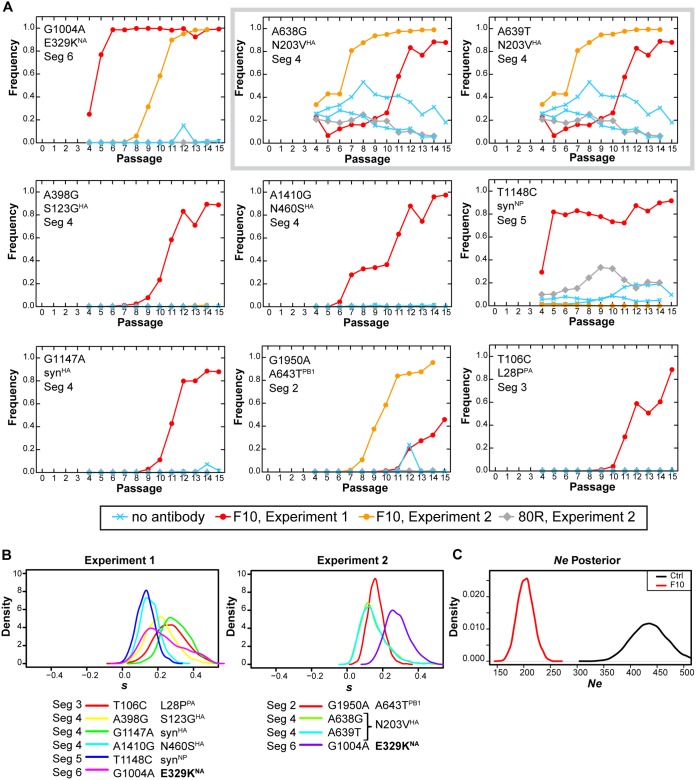
FIG 2.
Mutations inferred to be evolving under positive selection in the presence of the broadly neutralizing antibody F10. (A) Trajectories of select mutations elicited by viral passaging with F10, with 80R control antibody, or without antibody, in terms of allele frequency. Mutations individually marked as A638G and A639T (gray box) are in perfect linkage and yield N203V, as the wild-type sequence is GGT AAC CAA (AAC, positions 638/639/640; protein, GNQ). The mutant sequence is GGT GTC CAA (GTC, positions 638/639/640; protein, GVQ). (B) The posterior probability distribution of selection coefficients (s) for the mutations for experiments 1 and 2. Specific mutations are listed by influenza viral protein, nucleotide change, and amino acid change. Seg, segment; Syn, synonymous. (C) Posterior distributions of effective population size inferred from WFABC. The effective population size was estimated from time-sampled genomic data assuming neutrality. For F10-treated (F10) and control (ctrl) samples, we respectively estimated Ne to be 208 (99% highest posterior density interval, 162, 249) and 440 (99% highest posterior density interval, 350, 512).