Figure 3. Schematic representation of de novo peroxisome biogenesis pathways in yeast and mammals.
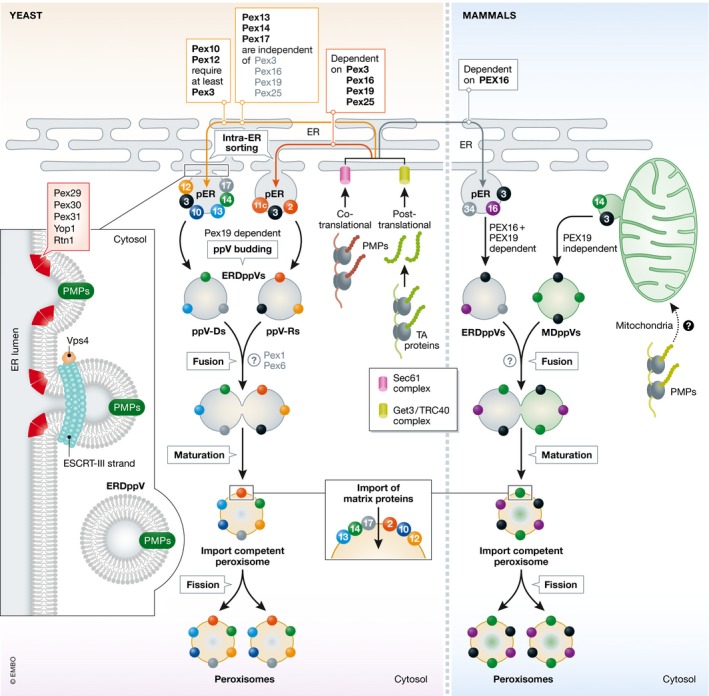
The first step in the de novo biogenesis is the indirect import of PMPs to the ER. Some PMPs are co‐translationally inserted into the ER membrane via the Sec61 complex 33, 108 and TA PMPs are post‐translationally incorporated into the ER membrane via the Get3 complex (yeast) 63 or ASNA1/TRC40 (mammals) 62. After PMP insertion into the ER, work in yeasts shows that an intra‐ER sorting step targets the PMPs to sub‐domains of the ER called the pER 32. Work in P. pastoris reveals that this routing of PMPs is either dependent or independent of Pex3, 16, 19, and 25 116, 117, 118. Studies from several yeasts define at least two modes of intra‐ER sorting of PMPs. One pathway is exemplified by the docking subcomplex proteins (Pex13, 14, and 17), which are independent of Pex3, 16, 19, and 25 118, 137. The other is exemplified by the RING‐domain PMPs (Pex10, 12, 2, 11c) and is dependent on Pex3, 16, 19, and 25 for intra‐ER sorting 118, 137. The exit sites for ppV budding are marked by the presence of several proteins (shown in inset on the left) including Pex29, 30, 31, which interact with Yop1 and Rtn1 and impart positive curvature in the ER 122, 124, 130. Subsequently, ESCRT‐III proteins (Vps20 and Snf7) are proposed to play a role in ppV scission 121 in an energy‐dependent manner, perhaps facilitated by Vps4 (stimulating disassembly of ESCRT‐III at the ER) 121. The ppVs bud from the pER in a Pex19‐dependent manner 115, 120. ERDppVs are of two distinct varieties—ppV‐R, containing Pex3, Pex2, and Pex11C and ppV‐D comprised of Pex13, Pex14, Pex17, Pex10, Pex12, and Pex3 116. Subsequently, these ppVs fuse heterotypically or with pre‐existing peroxisomes 40, 42, 119. In mammals, ppV formation is different in that several PMPs are sorted to the pER in a PEX16‐dependent manner 31, 146 and several other PMPs are routed to peroxisomes via mitochondria, from which MDppVs are formed in a PEX19‐independent manner 39. Subsequently, ERDppVs and MDppVs are proposed to fuse to form import‐competent peroxisomes, which subsequently import the matrix proteins and become metabolically active organelles. The question mark (?) represents uncertainty regarding either the known 42, 119, 144 or unknown proteins required for this fusion step.
