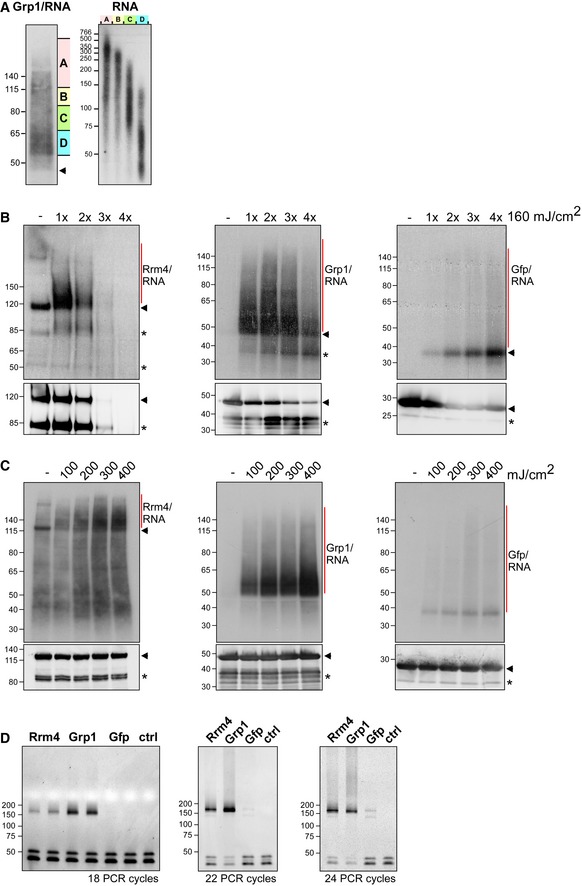
Grp1‐Gfp/RNA complexes were size‐separated on denaturing PAGE after UV‐C irradiation and transferred to a nitrocellulose membrane (left). RNA was radioactively labelled, and protein‐RNA complexes with covalently linked RNAs of different sizes were visible as smear above the expected molecular weight of the Grp1‐Gfp protein (45 kDa; marked by arrowhead). RNA of four different regions of the membrane (A–D indicated on the right) were isolated from the membrane and were size‐separated on a denaturing gel (6%) (right; nucleotide size marker on the left, bp).
Autoradiographs showing Rrm4‐Gfp, Grp1‐Gfp and Gfp in complex with RNA after UV‐C irradiation at 0, 160, 320, 480 and 640 mJ/cm2. Corresponding Western blots using anti‐Gfp are shown below. Arrowheads indicate the expected molecular weight of the proteins (Rrm4‐Gfp, 112 kDa; Grp1‐Gfp, 45 kDa; Gfp, 27 kDa). After each irradiation step, the cells were mixed. Note that increased UV‐C irradiation in combination with slow processing due to long time intervals was particularly harmful to the Rrm4 protein, which was completely degraded after four minutes of UV‐C irradiation. Putative degradation products are marked by asterisks.
Autoradiographs showing Rrm4‐Gfp, Grp1‐Gfp and Gfp in complex with RNA after single UV‐C irradiation at 0, 100, 200, 300 or 400 mJ/cm². This time, mixing breaks were omitted and cells were harvested as quickly as possible. Corresponding Western blots are shown below. Labelling as above. We chose 200 mJ/cm2 as optimal UV‐C irradiation dose, since the amount of unspecific Gfp‐RNA complexes increased at higher doses. Arrowheads and asterisks as in (B).
Amplification of the Rrm4‐, Grp1‐ and Gfp‐derived cDNA libraries with different numbers of PCR cycles (between 18 and 24; ctrl, control without template cDNA). The PCR products were separated on a native gel (6%) and stained with SYBR green I (nucleotide size marker on the left, bp). The size of the cDNA insert together with the adapters (cDNA insert = 20–30 nt; L3 adapter, RT‐primer and P3/P5 Solexa primers = 128 nt) is expected to be ˜ 150–160 nt after amplification.