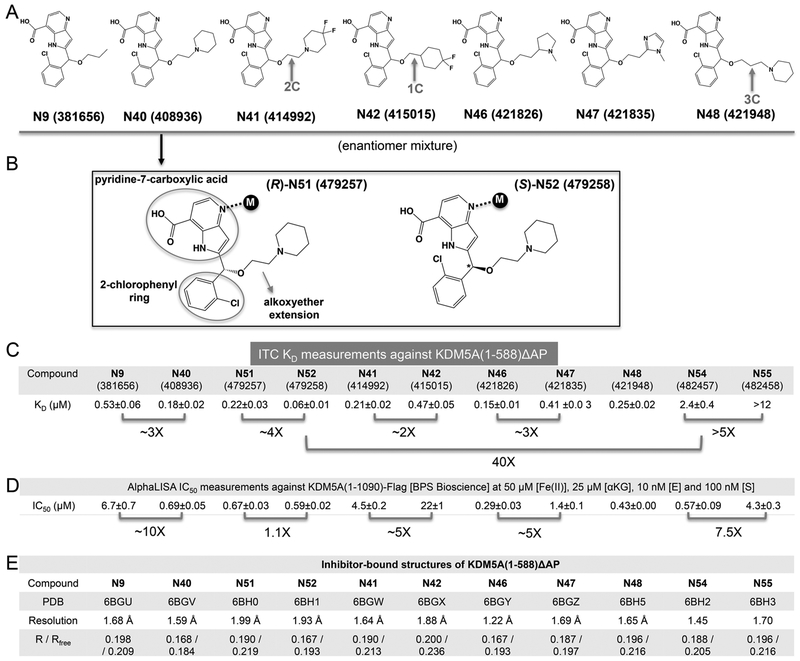
Figure 1.
Binding of KDM5A by compound N9 and its derivatives. (A) Chemical structures of N9 related compounds containing a constant pyridine-7-carboxylic acid moiety and 2-chlorophenyl moiety connected to a variable hydroxymethyl extension. (B) Chemical structures of (R)- and (S)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (compounds N51 and N52). (C) Summary of isothermal titration calorimetry measurement of dissociation constants (KD) of various compounds to KDM5A(1–588)ΔAP (see Figure S2 for original binding data). (D) Summary of the IC50 values of inhibition of demethylation for KDM5A (residues 1–1090) by AlphaLISA (see Figure S3 for original inhibition data). (E) Summary of inhibitor-bound X-ray structures of KDM5A(1–588)ΔAP (see Table S1 for detailed statistics of X-ray diffraction and refinement).