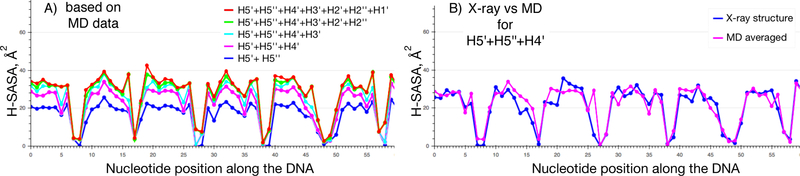
Figure 5. H-SASA profiles for DNA in nucleosome.
A) H-SASA (solvent accessible surface area of deoxyribose hydrogen atoms) profiles are calculated using contributions from different deoxyribose hydrogen atoms for each nucleotide along the DNA sequence starting from the center of the nucleosome. Profiles were calculated by averaging over 50 snapshots from MD simulations spaced 1 ns apart. B) Comparison of H-SASA profiles (based on H5’, H5’’ and H4’ atoms) as calculated from X-ray structure (with hydrogen atoms added via REDUCE) and MD trajectory. If only H5’, H5’’ and H4’ atoms are available, profiles do not differ much and X-ray structures can be used for reliable profile estimation. Zero on the x-axis marks the nucleotide positioned on the central dyad symmetry axis of nucleosome.