Fig. 6.
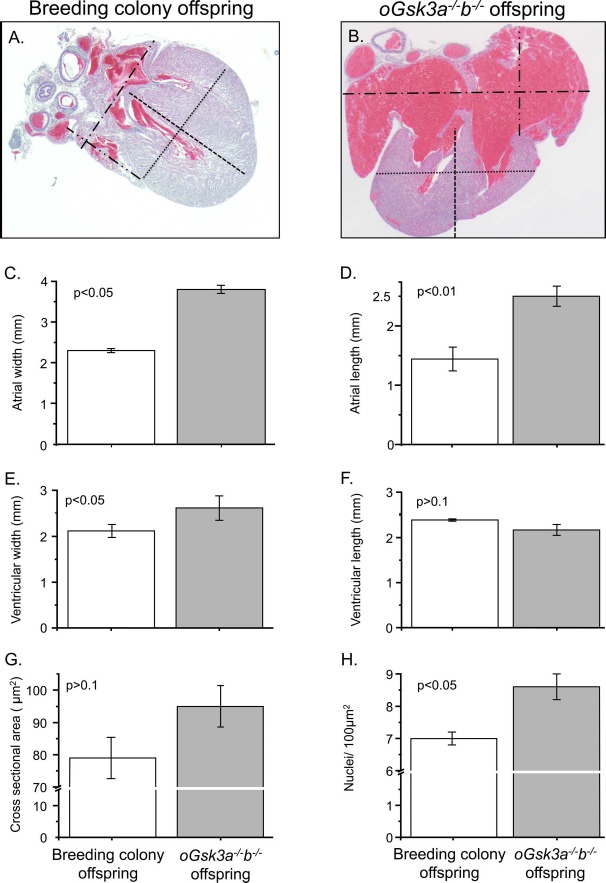
Comparison of the breeding colony and oGsk3a−/−b−/− offspring heart phenotypes. A and B) Representative micrographs used to quantify atrial and ventricular dimensions of hearts from mouse breeding colony and oGsk3a−/−b−/− female-derived offspring (×25 magnification): atrial width, dash-dotted lines; atrial length, dash-dot-dotted lines; ventricular width, dotted lines; and ventricular length, dashed lines. B) Atria from oGsk3a−/−b−/− female-derived pups were visually increased in relation to atria from mouse breeding colony offspring. C and D) Atrial width and length were significantly greater in oGsk3a−/−b−/− offspring hearts (n = 6) in comparison to atrial dimensions of mouse breeding colony offspring hearts (n = 3). E) Ventricular width was greater in hearts from oGsk3a−/−b−/− offspring (n = 6) in relation to mouse breeding colony offspring hearts (n = 3); however, the ventricular length was similar between groups (F). G) Cross-sectional area of cardiomyocytes in heart ventricles from mouse breeding colony (n = 5) and oGsk3a−/−b−/− (n = 5) offspring were similar, but apexes from oGsk3a−/−b−/− female-derived animals had significantly greater cell density than in mouse breeding colony offspring hearts (H). Values in column graphs are means ± SEM.
