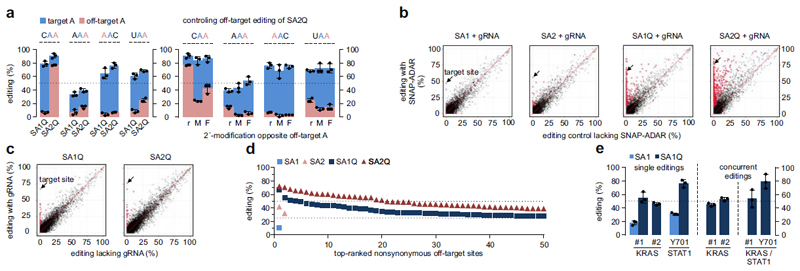
Figure 2. Editing specificity and application.
a) Off-target editing of adjacent adenosines in A-rich triplets. r, M, F refers to the chemical modification opposite of the off-target A with r = natural ribonucleotide, M = 2´-methoxy, F = 2´-fluoro. b), c) Scatter plots of differential editing at ≈50.000 sites/experiment. The targeted site (ACTB) is indicated by an arrow. Significantly differently edited sites (p<0.01) are highlighted as red dots. In b), editing is compared to a control cell line lacking SA expression. c) shows editing in presence versus absence of the gRNA. d) Nonsynonymous off-target sites ranked by editing yields. e) Editing of signaling transcripts. Two 5´-UAG sites in the ORF of KRAS (#1, #2), and a 5´-UAU site in STAT1 (Tyr701) were targeted. For concurrent editing, two respective gRNAs were cotransfected into SA1Q cells. Data in a), e) are shown as the mean±SD, N=3 independent experiments, black dots represent individual data points. Significance in b), c) was tested by Fisher´s exact test (two-sided), N=2 independent experiments.