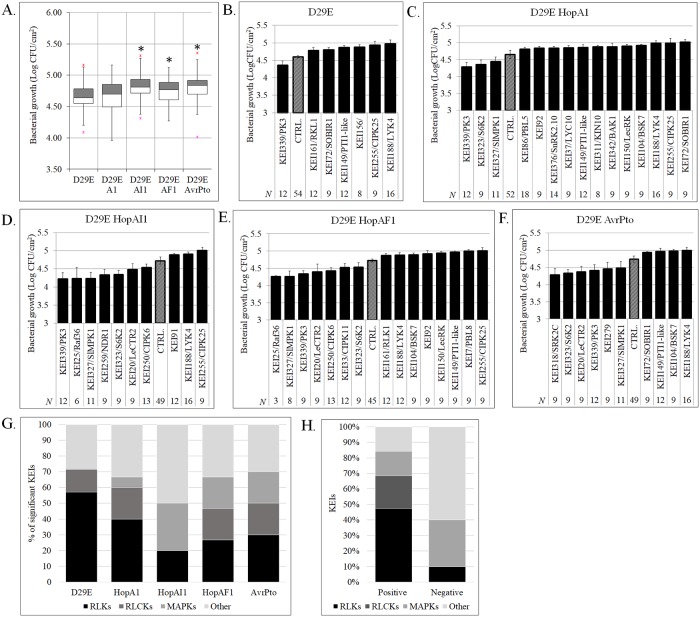
Fig 3. Plant kinases promote defense or susceptibility to Pseudomonas syringae.
A. Quantification of P. syringae growth without effectors (D29E) or with one effector (D29E + HopA1, D29E + HopAI1, D29E + HopAF1, or D29E + AvrPto) in Nicotiana benthamiana leaves transformed with the EC1 control plasmid. Statistically significant events (*) are considered at p < 0.01 compared with D29E. B–F. Quantification of the growth of P. syringae strains in N. benthamiana leaves silenced for individual kinases (KEIs), where only KEIs with statistically significant (p < 0.05) phenotypes were plotted. The control values (CTRL.) represent quantification of the P. syringae growth in plants silenced by the EC1 control. G. Histogram with the percentage and structural class of KEIs identified as regulators of pathogen growth in plant tissues inoculated with the P. syringae strains D29E, D29E + HopA1, D29E + HopAI1, D29E + HopAF1, and D29E + AvrPto. H. Histogram with the number of KEIs shown to act as candidate positive or negative regulators of immunity based on all infection assays. CFU, colony-forming unit; KEI, Kinase Effector Interactor; MAPK, MAP kinase; N, number of independent replicates; RLCK, receptor-like cytosolic kinase; RLK, receptor-like kinase.