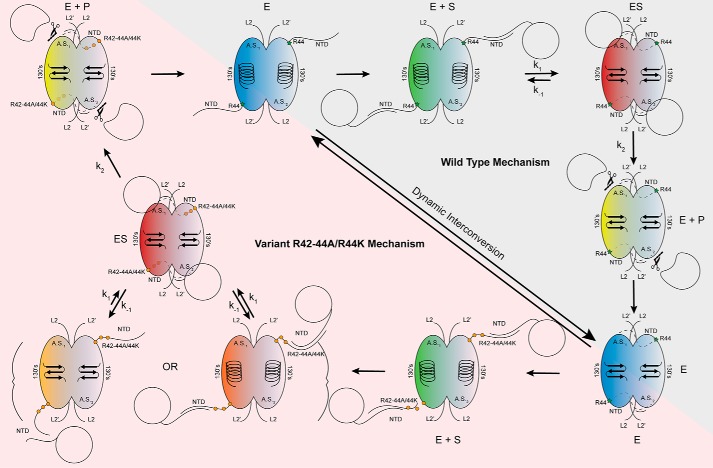
Figure 10.
Model of caspase-6 protein substrate recognition. In the mature state, caspase-6 dynamically interconverts between the unique helical conformation and the canonical strand conformations (blue) (30). Herein, we propose, consistent with other models, that the intrinsically disordered NTD of caspase-6 fishes for protein substrates within the context of the cell, utilizing the unique sequences and charges of the domain to tune protein substrate recognition (green). The intrinsically disordered domain of caspase-6 is likely binding to the disordered domains of the substrate proteins (21, 55). The NTD orients the protein substrate, facilitating formation of the enzyme–substrate complex, by docking with the protein-binding interface of caspase-6. This step also coincides with the transition to the obligate strand state, which is required for enzymatic processing (red). Once assembled, chemistry proceeds producing cleaved product (yellow), wherein the remaining substrate is released allowing transition back to the dynamic resting state of mature caspase-6 (blue). Substitutions of the exosite at 42RRR44 are not proposed to affect the dynamic resting state of caspase-6, as it will still undergo helix-to-strand transitions, albeit at a different rate (blue) (Fig. 7A). However, upon substrate recruitment (green) and transition into the ES complex (red), the substitutions at the exosite of caspase-6 impact the dynamics of the NTD (Fig. 7), unhinging it, leading to perturbations of the protein docking face of the enzyme (Fig. 9, B and C) and the decreased rate of full substrate engagement, observed indirectly through the increased KD apparent of substrate binding (orange) (Fig. 9, D–F). Once the ES complex is successfully assembled (red), chemistry will proceed as expected (Fig. 4, A and B) leading to product formation (yellow) and back to the resting state of caspase-6 (blue).