Figure 9.
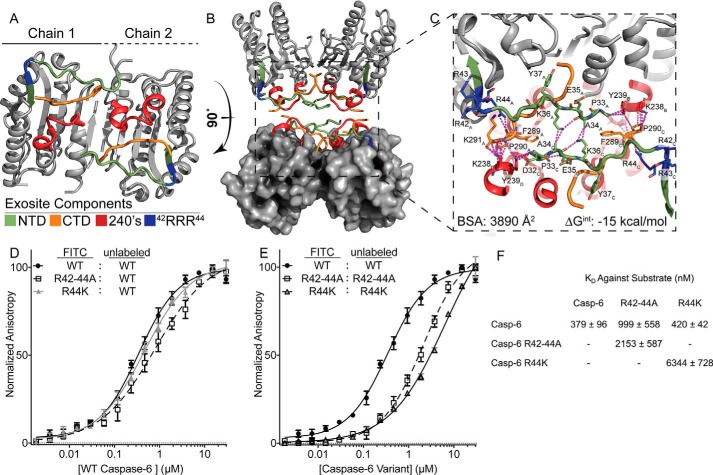
Exosite is critical for stabilization of putative protein-binding interface. A, regions identified by H/Dx-MS are displayed on the caspase-6 structure 3K7E. The NTD (green) containing the exosite patch (blue) and 240's regions (red) make contacts with the CTD (orange) on the reverse face of caspase-6. B, rotating caspase-6 90° reveals a potential binding interface that occurs in the tetrameric helical structures of caspase-6, incorporating all regions identified by H/Dx. C, putative binding interface analyzed by PISA (58), containing large portions of buried surface area primarily consisting of buried hydrophobics. Residue label subscripts correspond to the chain of the crystal structure. D, fluorescence polarization assay of FITC-labeled caspase-6 C163S FL constructs of either the WT (black), R42–44A (white), or R44K (gray) variants incubated with the WT unlabeled protein as a substrate to assess KD apparent. E as in D except incubated with their respective unlabeled R42–44A C163S FL or R44K C163S FL variants. F, KD values of all labeled proteins incubated with the unlabeled WT substrate are not statistically significantly different, as compared to those incubated with R42–44A or R44K unlabeled proteins, which have a weaker KD value. In all cases, the caspase-6 variants are the ΔN D179CT form.