Abstract
SMARCC2 (BAF170) is one of the invariable core subunits of the ATP-dependent chromatin remodeling BAF (BRG1-associated factor) complex and plays a crucial role in embryogenesis and corticogenesis. Pathogenic variants in genes encoding other components of the BAF complex have been associated with intellectual disability syndromes. Despite its significant biological role, variants in SMARCC2 have not been directly associated with human disease previously. Using whole-exome sequencing and a web-based gene-matching program, we identified 15 individuals with variable degrees of neurodevelopmental delay and growth retardation harboring one of 13 heterozygous variants in SMARCC2, most of them novel and proven de novo. The clinical presentation overlaps with intellectual disability syndromes associated with other BAF subunits, such as Coffin-Siris and Nicolaides-Baraitser syndromes and includes prominent speech impairment, hypotonia, feeding difficulties, behavioral abnormalities, and dysmorphic features such as hypertrichosis, thick eyebrows, thin upper lip vermilion, and upturned nose. Nine out of the fifteen individuals harbor variants in the highly conserved SMARCC2 DNA-interacting domains (SANT and SWIRM) and present with a more severe phenotype. Two of these individuals present cardiac abnormalities. Transcriptomic analysis of fibroblasts from affected individuals highlights a group of differentially expressed genes with possible roles in regulation of neuronal development and function, namely H19, SCRG1, RELN, and CACNB4. Our findings suggest a novel SMARCC2-related syndrome that overlaps with neurodevelopmental disorders associated with variants in BAF-complex subunits.
Keywords: developmental delay, intellectual disability, neurodevelopmental disorder, Bafopathy, transcriptome, genotype-phenotype correlation, speech delay, dysmorphisms
Main Text
The chromatin-remodeling complex BRG1-associated factor (BAF) plays an essential role in the regulation of gene expression and higher-order chromatin organization by modulating the nucleosome and changing chromatin conformation and accessibility.1, 2 BAFopathies are a heterogeneous group of disorders caused by mutations in the various subunits composing the BAF complex. The clinical phenotypic spectrum of BAFopathies is wide and involves various human neurodevelopmental disorders including syndromic and nonsyndromic intellectual disability (ID), growth retardation,3, 4, 5, 6, 7, 8, 9, 10 sporadic autism,11 schizophrenia,12 and amyotrophic lateral sclerosis.13
The most recognizable syndrome associated with BAF abnormalities is Coffin-Siris syndrome (CSS [MIM: 135900]). This is a genetically heterogeneous ID syndrome characterized by developmental delay (DD), speech delay, coarse facial appearance, feeding difficulties, and hypoplastic-to-absent fifth finger nails and fifth distal phalanges.14 This syndrome is associated with abnormalities in multiple subunits of the BAF complex including the ATPase subunit SMARCA4 (MIM: 603254), the common core subunit SMARCB1 (MIM: 601607), and BAF accessory subunits such as SMARCE1/BAF57 (MIM: 603111), ARID1A (MIM: 603024), ARID1B (MIM: 614556),15 ARID2 (MIM: 609539),16 and DPF2 (MIM: 601671).17 CSS can result of pathogenic changes in other chromatin remodeling proteins with no direct interaction with BAF complex, including SOX11 (MIM: 600898)18 and PHF6 (MIM: 300414).19
Other BAFopathies include Nicolaides-Baraitser syndrome (MIM: 601358) caused by pathogenic variants in SMARCA2 (MIM: 600014) that has a significant phenotypic overlap with CSS and is characterized by ID, sparse hair, short stature, microcephaly, brachydactyly, interphalangeal joint swellings, and epilepsy.20, 21 Some individuals with clinical diagnosis of DOORS syndrome (MIM: 220500), characterized by deafness, onychodystrophy, osteodystrophy, ID, and seizures, were found to carry pathogenic variants in SMARCB1 (MIM: 601607), highlighting the clinical overlap between CSS and DOORS syndrome.22 We have previously reported mutations in another BAF subunit, ACTL6A (MIM: 604958), to be associated with ID.23 Pathogenic variants in ADNP (MIM: 611386), encoding a transcription factor that interacts with the BAF complex, have been identified in individuals presenting dysmorphic facial features, autism spectrum disorder (ASD), ID, hypotonia, and congenital heart defects.24 Detailed phenotypic and genetic comparison between the different BAF-related syndromes has been discussed elsewhere.4, 25
SMARCC2 (MIM: 601734) encodes BAF170, a common core subunit of the BAF complexes with high homology to SMARCC1 (BAF155).26 It is an intrinsic factor of glial radial cells and plays a crucial role in embryogenesis and corticogenesis, determining the mammalian body and cortical size.27 Smarcc2;Smarcc1 double knockout mice demonstrated proteasome-mediated degradation of the entire BAF complexes, resulting in impairment of the global epigenetic and gene expression program of forebrain development.28 Recently, deletion of Smarcc2 in mice revealed its role in learning and behavioral adaptation.29 SMARCC2 was also reported as one of the chromatin-remodeling genes involved in ASD.30 Despite its significant biological role, variants in SMARCC2 have not been directly associated with a syndrome in humans previously.
We report 15 unrelated individuals (Tables 1 and S1) with variants in SMARCC2, detected by whole-exome sequencing (WES), and with clinical presentation that includes mild to severe ID (HP:0012736), DD with prominent speech delay (HP:0000750), behavioral abnormalities (HP:0000708), growth retardation (HP:0008897), feeding difficulties at the neonatal period (HP:0008872), hypotonia (HP:0011398), and dysmorphic features including hypertrichosis (HP:0000998), thick eyebrows (HP:0000574)/prominent supra-orbital ridges (HP:0000336), and thin upper and lower vermillion (HP:0000233), suggesting overlap with Coffin-Siris and Nicolaides-Baraitser syndromes. Subjects in this cohort were gathered using GeneMatcher.31 All individuals’ families from the different institutions agreed to participate in this study and signed appropriate consent forms. Permission for clinical photographs was given separately. Individual 4 has been reported before and was identified in a gene panel screening (1,256 genes) of 96 individuals with ID.32 The variant c.1833+2T>C in individual 10 was reported before as part of a work to identify new gene-disease associations in trio WES from 119 undiagnosed individuals.33 Individuals 1, 4, 6, 7, 9–13, and 15 had trio WES. Individuals 2, 3, 5, and 14 had a proband WES followed by Sanger confirmation for the individual and parents. Biological parents of individuals 3 and father of individual 8 are not available for testing. Individual 8 had duo WES with his mother and results were re-analyzed as part of the Undiagnosed Diseases Network (UDN). This individual was found to harbor an intronic splice site variant c.1833+1G>T in SMARCC2 that was not found in his mother. Twelve of the fifteen individuals have proven de novo variants in SMARCC2. Individual 2 inherited the variant from his affected father. Paternal grandparents were negative for the variant indicating a de novo variant in the father.
Table 1.
Summary of SMARCC2 Variants and Clinical Presentation of 15 Individuals
| Individual # | # 1 | # 2 | # 3 | # 4a | # 5 | # 6 | # 7 | # 8 | # 9 | # 10b | # 11 | # 12 | # 13 | # 14 | # 15 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | M | M | M | F | M | F | M | M | M | M | M | F | M | M | M | |
| Age (Y) | 5 Y | 3 Y | 2 Y | 17 Y | 4 Y | 18 Y | 11 Y | 11 Y | 2.5 Y | 10.5 Y | 19 Y | 7 Y | 8 Y | 5 Y | 27 Mo | |
| Nucleotide change | c.400A>G | c.723G>A | c.999dupA | c.1311−3C>G | c.1826T>C | c.1829T>C | c.1829T>C | c.1833+1G>T | c.1833+2T>C | c.1833+2T>C | c.1838T>C | c.1903T>C | c.2686A>G | c.2699A>G | c.3456_3458delCAT | |
| Amino acid change | p.Asn134Asp | p.Trp241∗ | p.Glu334Argfs∗49 | splicing variant | p.Leu609Pro | p.Leu610Pro | p.Leu610Pro | splicing variantc | splicing variant | splicing variant | p.Leu613Pro | p.Cys635Arg | p.Met896Val | p.Glu900Gly | p.Met1153del | |
| De novo variant? | yes | no (affected father) | n/a | yes | yes | yes | yes | n/a | yes | yes | yes | yes | yes | yes | yes | |
| Affected domain | SMARCC_N | SMARCC_N | SMARCC_N | SWIRM | SANT | SANT | SANT | SANT | SANT | SANT | SANT | SANT | SMARCC_C | SMARCC_C | carboxy terminal | |
| Neurodevelopmental Abnormalities | ||||||||||||||||
| Developmental delay and/or intellectual disability | mild DD | mild DD | mild DD | severe DD | severe DD | severe DD | severe DD | severe DD (DQ=20) | moderate ID | moderate DD, moderate-severe ID | moderate-severe DD | moderate DD, moderate ID | moderate-severe DD | mild ID | mild DD | 15/15 |
| Speech impairment | − | speech delay | − | absence of language | absence of language | absence of language | absence of language | absence of language | absence of language | minimal speech | absence of language | speech delay | speech delay | speech delay | speech abnormalities | 13/15 |
| Behavioral abnormalities | − | + | + | + | + | − | + | + | − | − | − | + | + | + | + | 10/15 |
| Hypotonia | + | spasticity | + | + | + and spasticity | + | + | + | + | + | − | + | + | + | + | 13/15 |
| Seizures | − | − | − | + | + | − | + | − | − | − | + | − | − | − | − | 4/15 |
| Movement disorder | − | + | − | − | − | + | − | − | − | − | − | − | − | − | − | 2/15 |
| CNS abnormalities on MRI | n/a | two discrete hyperintens white matter lesions | n/a | normal MRI | thinning of corpus callosum and splenium, periventricular white matter loss | generalized cerebral atrophy, hypointensity globus pallidus | generalized cerebral atrophy, hypo-myelination | normal MRI | normal MRI | normal MRI | abnormal corpus callosum | slightly small corpus callosum, prominent perivascular spaces | normal MRI | normal MRI | n/a | 6/12 |
| Growth | ||||||||||||||||
| FTT | − | − | − | − | + | + | + | − | + | + | − | − | − | + | − | 6/15 |
| Sucking/feeding difficulty | − | − | − | − | + | + | + | − | + | + | + | − | − | + | + | 8/15 |
| Craniofacial Features | 11/15 | |||||||||||||||
| Thin/sparse scalp hair | − | − | − | − | + | + | − | − | + | − | + | − | − | − | − | 4/15 |
| Hypertrichosis | − | − | − | + | + | − | − | − | + | − | − | + | + | + | − | 6/15 |
| Thick eyebrows | − | − | − | + | − | + | + | − | + | − | + | − | − | + | − | 6/15 |
| Long eyelashes | − | − | − | − | − | − | − | − | + | + | + | − | + | + | − | 5/15 |
| Ptosis | − | − | − | − | − | + | + | − | − | + | + | − | − | + | − | 5/15 |
| Thin upper lip vermilion | − | − | − | − | + | + | − | + | + | − | − | + | + | − | − | 6/15 |
| Thick lower lip vermilion | − | − | − | + | − | − | + | − | + | − | + | − | + | − | − | 5/15 |
| Palate abnormalities | − | − | − | − | − | − | − | − | − | + | + | − | − | + | − | 3/15 |
| Nose upturned/anteverted nostrils | − | − | − | − | + | + | + | − | − | + | + | + | − | − | − | 6/15 |
| Skeletal-Limb Features | ||||||||||||||||
| 5th finger or toe/nails abnormalities | − | − | − | − | + | − | − | − | − | + | + | − | − | + | − | 4/15 |
| Scoliosis | − | − | − | kyphosis | + | + | + | − | − | − | − | + | − | − | − | 5/15 |
| Other | ||||||||||||||||
| Cardiovascular | n/a | − | − | − | n/a | left coronary distension | non-progressive mild aortic dilatation (Z score=2.54). | − | − | − | − | − | n/a | − | n/a | 2/11 |
| Inguinal hernia | − | − | − | − | + | − | + | + | − | − | − | − | − | − | − | 3/15 |
| Undescended testis | n/a | − | − | n/ap | + | n/ap | + | − | n/a | − | − | n/ap | − | − | n/a | 2/12 |
| Skin problems | − | hypo pigmented hair, cafe au lait macules | − | − | eczema | eczema, scleroderma | − | − | hypo-melanotic macula | vitiligo (present in unaffected father) | hyper pigmented irregular skin on back | eczema | − | − | − | 7/15 |
Abbreviations: FTT, failure to thrive; DD, developmental delay; ID, intellectual disability; DQ, developmental quotient; CNS, central nervous system; MRI, magnetic resonance imaging; M, male; F, female; Y, years; Mo, months; n/a, not available information; n/ap, not applicable. Minus sign (−), not reported in this individual; plus sign (+), reported in this individual.
See Martínez et al.32
See Zhu et al.33
See Figure 3.
All of the individuals presented here have some degree of ID and/or DD (Tables 1 and S1). Ten (10/15, 65%) have moderate to profound DD and ID while the other five individuals have only mild ID or mild DD. 13/15 (86%) individuals have speech impairment with 7 of them completely lacking language. Most individuals present muscle tone abnormalities. 13/15 (85%) individuals present significant hypotonia, while two of the individuals present high tone or spasticity. Ten of the individuals (67%) present behavioral problems including aggression and self-injurious behavior as well as hyperactivity, hypersensitivity to touch, sleep disturbances, and obsessive and rigid behavior. Two were noted to have difficulties in social interactions, yet not qualified for formal diagnosis of autism. Eight individuals present feeding difficulties and six of them have mostly postnatal growth retardation. Individual 9 has continuous feeding difficulties with laryngomalacia and nasal feeding tube since age 14 months. 11 out of the 15 individuals are reported to have dysmorphic craniofacial features (Figure 1). The most pronounced dysmorphic features are hypertrichosis (6/15), thick eyebrows/prominent supra-orbital ridges (6/15), thin upper or thick lower lip-vermilion (6/15 and 5/15, respectively), and upturned nose (6/15). Most of the individuals have normal fifth finger/toe and finger/toenail.
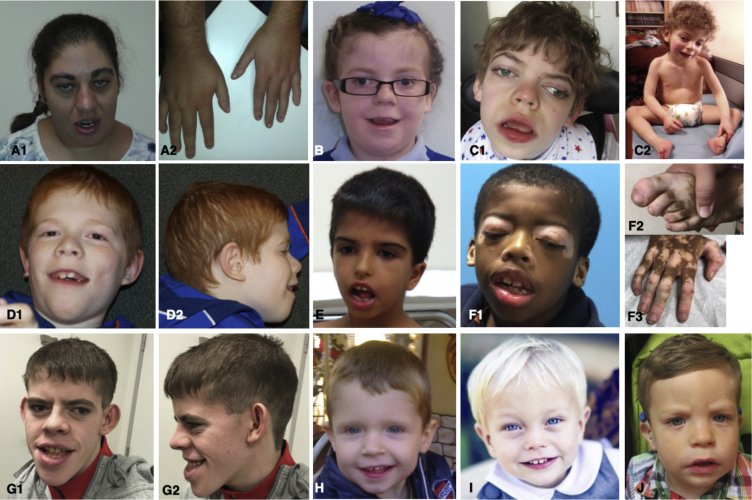
Figure 1.
Pictures of Ten of the Individuals with SMARCC2 Variants
(A) Individual 4 (c.1311−3C>G) at 17 years of age.
(B) Individual 12 (p.Cys635Arg) at 7 years of age.
(C) Individual 7 (p.Leu610Pro) at 11 years of age.
(D) Individual 8 (c.1833+1G>T) at 11 years.
(E) Individual 14 (p.Glu900Gly) at 5 years of age.
(F) Individual 10 (c.1833+2T>C) at 10.5 years of age (father of this individual presents vitiligo as well).
(G) Individual 11 (p.Leu613Pro) at 19 years of age.
(H) Individual 2 (p.Trp241∗) at 3 years of age.
(I) Individual 3 (p.Glu334Argfs∗49) at 2 years of age.
(J) Individual 15 (p.Met1153del) at 2 years of age.
Note coarse facial features (A, C, G), thick eyebrows/prominent supra-orbital ridges, long eyelashes, upturned nose, open mouth with thin upper lip vermilion, and hypertrichosis. Note lateral commissures creating skin indentation seen among most affected individuals. Note camptodactyly of 5th fingers and toes (F2, F3).
8 of the 15 individuals presented here (1, 5–7, 11–14) have one of seven missense variants in SMARCC2 (GeneBank: NM_003075.3) and all are predicted to be deleterious/probably damaging according to PROVEAN and SIFT, in silico tools to predict the functional effect of an amino acid substitution. All missense variants are in well-conserved amino acids in SMARCC2 (Figure 2). In vitro missense tolerance ratio (MTR) tool shows that six of the seven missense variants presented here preferentially affect one of the 25% most intolerant residues of SMARCC2 (p = 0.0004) with p.Asn134Asp being the only missense variant affecting a tolerant region of this gene (Figure S1). This might be consistent with the milder phenotype presented in this individual. SMARCC2 ExAC z-score for intolerance for missense variations is significantly high (4.26) and the gene is predicted to be potentially associated with dominant conditions according to an LDA score of 2.435 by the DOMINO algorithm.34 Individual 15 harbors a de novo in-frame deletion of methionine 1153 (c.3456_3458delCAT). Methionine 1153 deletion and its missense substitutions to various amino acids (isoleucine, threonine, valine) appear once (allele frequency 3.231 × 10−5) and four times, respectively, in gnomAD.35 This might indicate that this position is relatively tolerable for changes and explain the mild clinical presentation of individual 15. It is important to note that detailed phenotype such as IQ scores are not available for individuals included in the ExAC database.
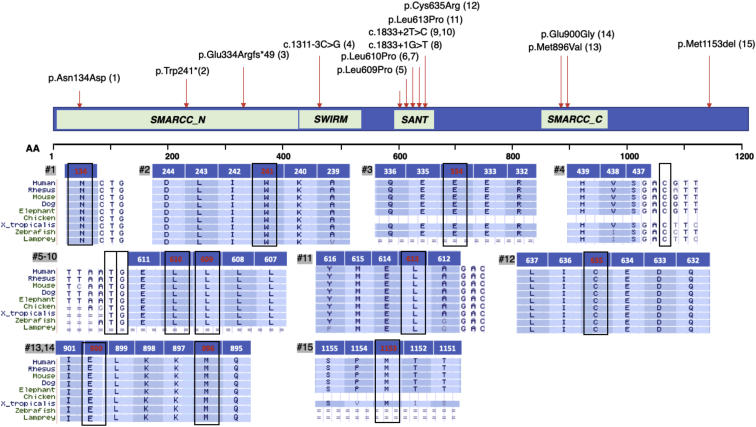
Figure 2.
SMARCC2 (BAF170) Main Domains and Variants
Amino acid alignments demonstrate that the missense variants affect highly conserved amino acids and the variants causing splicing abnormalities affect highly conserved nucleotides (GenBank: NP_003066, transcript ENST00000267064.8). The involved amino acids/nucleotides are framed in black. Number of relevant individual (#) is on gray background. For intron/exon location of the different variants, see Table S1.
Individuals 2 and 3 carry truncating mutations located at the N terminus of SMARCC2 (exons 9 and 11). Individual 2 carries a heterozygous nonsense variant (p.Trp241∗) and individual 3 carries a frameshift variant (p.Glu334Argfs∗49). Interestingly, both present mild phenotype. Individual 2, who presents mainly behavioral abnormalities and only mild DD, inherited the change from his affected father who presents with borderline intelligence (IQ of 72) and behavior problems. ExAC database identified only a single loss-of-function allele in SMARCC2 (p.Glu389Aspfs∗5); however, with only 11 (22%) of 49 reads at this frameshift variant site, it is unlikely to be a germline variant (binomial exact test for 50% germline heterozygous expectation; p = 0.0001). The depletion of protein-truncating variants in SMARCC2 is evident in large human population reference cohorts. Based on the ExAC cohort, SMARCC2 achieves a LoF depletion FDR adjusted p = 5.5 × 10−11, ranking it among the 1.6% most significantly LoF-depleted genes in the human exome.36 It also has a probability of loss-of-function (LoF) intolerance (pLI) score of 1, supporting deleterious effect for predicted LoF pathogenic variants.35 SMARCC2%HI score of 20.29 also predicts that this gene is less likely to tolerate a loss-of-function variant or deletion.37
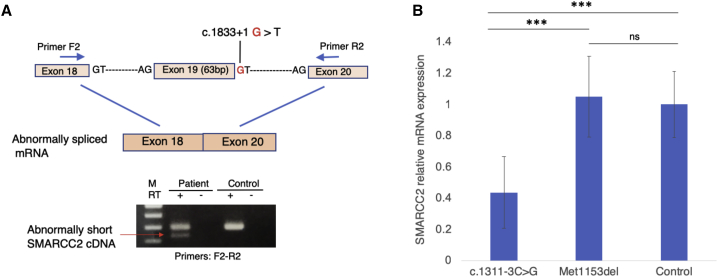
Four individuals harbor splicing variants. Three individuals (8–10) have one of two variants in intron 19 (c.1833+2T>C and c.1833+1G>T) that are predicted in silico to affect splicing donor site according to Human Splicing Finder (HSF). One individual (4) presents a splicing variant in intron 14 (c.1311−3C>G) that is predicted to affect exon acceptor site. In vitro assay in lymphocytes from individual 8 (c.1833+1G>T) indicates that this variant leads to abnormal splicing with deletion of exon 19 (amino acids 590–611) that compose the SANT domain in SMARCC2 (Figure 3A). The capture of a shorter cDNA product using RT-PCR might indicate the presence of an mRNA escaping nonsense-mediated decay (NMD). RT-qPCR quantification of SMARCC2 in lymphocytes from individual 4 (c.1311−3C>G) demonstrates a ∼50% decrease in mRNA compared to a control and a coding in-frame deletion (individual 15, p.Met1153del), suggesting haploinsufficiency through NMD (Figure 3B).
Figure 3.
Splicing Variants Analyses in Individuals 8 and 4
(A) PCR amplification of cDNA from individual 8 at exons 18 to 20 reveals deletion of exon 19 (amino acids 590–611) due to splicing variant c.1833+1G>T in SMARCC2 (GenBank: NM_003075).
(B) SMARCC2 gene expression was quantified by qRT-PCR from lymphoblastoid cell lines (LCLs) for individuals 4 and 15 and an unrelated control subject, in triplicates. SMARCC2 expression was normalized to GAPDH, and results are represented as relative expression normalized to the control. Error bars represent SEM (standard error of the mean) and significance was assessed with bilateral unpaired Student’s t tests.
Nine individuals carry missense or splicing variants in the highly conserved SWIRM (Swi3, Rsc8, and Moira) and SANT (Swi3, Ada2, NCoR, and TFIIIB) domains of SMARCC2 (Figure 2). The SWIRM domain interacts with DNA, binds di-nucleosome structures, and mediates specific protein-protein interactions.38 The SANT domain has DNA-binding activity39 and is believed to function as a histone tail binding module.40 Eight of the individuals reported here have a heterozygous novel variant in the SANT domain of SMARCC2. One individual has a missense variant changing leucine for proline in position 609 (p.Leu609Pro), two have a missense variant changing leucine for proline in position 610 (p.Leu610Pro), one individual has a missense variant changing leucine for proline in position 613 (p.Leu613Pro), and one has a missense variant changing cysteine to arginine in position 635 (p.Cys635Arg). Three individuals have one of two intronic variants that are predicted in silico to affect the same splicing donor site in SANT and one individual presents a splicing variant at the SWIRM domain. This group of nine individuals present with moderate to severe DD and ID and with severe speech impairment, with six of them having no language at all. Five out of these nine individuals (55%) have abnormal brain MRI findings including small corpus callosum and generalized cerebral atrophy. Five out of the nine present significant growth retardation and four of them have seizure disorder. It seems that individuals carrying a pathogenic variant in SWIRM or SANT domains have a more severe presentation. Interestingly, only the two individuals harboring the p.Leu610Pro variant have cardiovascular abnormalities. An echocardiogram for individual 6 revealed distended left coronary artery and for individual 7 mild non-progressive dilatation of the ascending aorta (Z score of 2.54).
To further investigate the pathogenicity of the variants, we calculated the geometric mean distance between the observed mutations to assay the clustering of the mutations.41 As this calculation considers only the length of the spliced transcript, intronic splicing mutations were annotated according to the closest coding nucleotide. Compared to ten million random permutations of mutations along the transcript, the observed mutations are significantly clustered (p value 10−7), suggesting a pathomechanism that does not operate through haploinsufficiency alone. Interestingly, removing the frameshift and nonsense mutations from the analysis does not affect the significance, but taking out the splicing mutations does (p value 0.12). This result points toward a more important role in the pathogenicity for the SWIRM and SANT domains than for the SMARCC_N region, and a corollary of this interpretation is the hypothesis that expressed but incorrectly spliced transcripts could function through a dominant-negative mechanism.
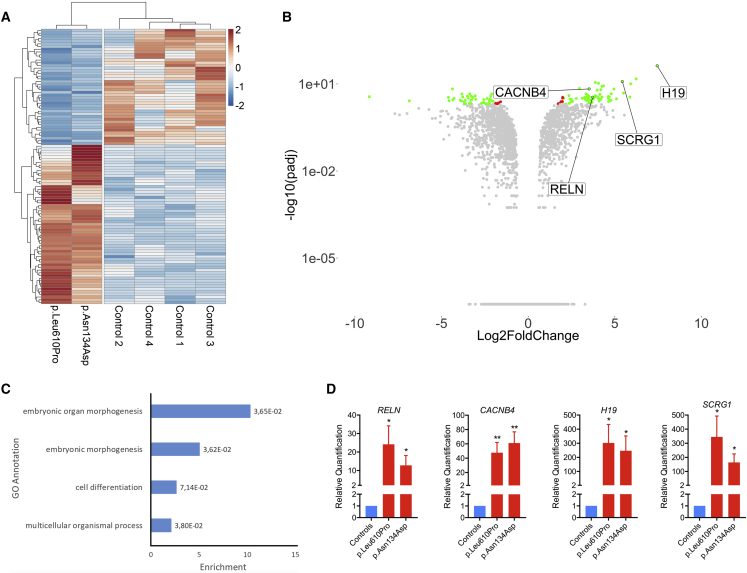
To better understand the impact of SMARCC2 mutations on gene expression, we performed an RNA-seq analysis on fibroblasts from individual 1 with p.Asn134Asp mutation and individual 7 with p.Leu610Pro mutation. We analyzed both samples together to assess shared differential gene expression patterns. Analysis of differentially expressed genes (DEGs) shows an almost equal amount of DEGs upregulated and downregulated in these two individuals (Figure 4A). Gene Ontology (GO) analysis of these DEGs using the GOrilla web application demonstrates a significant enrichment in genes related to embryonic morphogenesis, multicellular organismal process, and developmental process (Figure 4C). Among the shared DEGs between the two tested individuals, we identified genes with possible role in regulation of neuronal development and function, namely H19, SCRG1, RELN, and CACNB4.
Figure 4.
Fibroblasts Harboring p.Leu610Pro or p.Asn134Asp SMARCC2 Mutation Have Differential Gene Expression Patterns When Compared to Fibroblasts from Healthy Control Subjects
RNA-seq was performed on fibroblasts from four healthy controls and two affected individuals to assay gene expression.
(A) Heatmap of differentially expressed genes (DEGs). Log2 fold change (Log2FC) and p values adjusted for 10% false discovery rate (padj) were calculated for the two individuals and four control subjects, respectively pooled together using the “DESeq2” R package. Normalized count values from “DESeq2” were plotted and scaled row-wise using the “pheatmap” R package for genes with a padj lower than 0.01 and an absolute Log2FC value higher than 2.
(B) Volcano plot showing differentially regulated genes as fold-change versus adjusted p values. Log2FC and –log10(padj) values were plotted using the “ggplot2” R package as a volcano plot. Red dots represent genes with a padj lower than 0.01 and a abs(Log2FC) lower than 2. Green dots represent genes with a padj lower than 0.01 and a abs(log2FC) higher than 2.
(C) Histogram of the enriched GO annotations with FDR adjusted p values lower than 0.01 calculated using the GOrilla web application using DEGs for the two individuals pooled together.
(D) RT-qPCR analysis of mRNA expression profile for RELN, CACNB4, H19, and SCRG1 with p.Leu610Pro and p.Asn134Asp SMARCC2 mutations. The data are presented as “relative quantification” mean ± SEM from 6 independent control subjects (N = 2). A Student’s t test was used to determine the statistical significance. ∗p ≤ 0.05 versus control group; ∗∗p ≤ 0.01 versus control group.
H19 codes for a long noncoding RNA and is among the most upregulated DEGs (log2FC = 7.4, padj = 7.18 × 10−43) in both individuals compared to control subjects (Table S2). H19 is an imprinted gene and has been implicated in the regulation of growth and embryonic development, tumorigenesis, and epilepsy-induced astrocyte and microglia activation.42, 43, 44 Interestingly, knocking down SMARCA4 in MCF10A epithelial cell line also results in significant H19 upregulation1 suggesting that H19 expression is affected by BAF complex modification and that SMARCC2 p.Asn134Asp and p.Leu610Pro mutations could indeed affect the complex integrity and/or function.
SCRG1 is another significant DEG (log2FC = 5.43, padj = 9.30 × 10−13) mainly expressed in the human brain, and its level is highly regulated during postnatal development, being absent in the fetal brain.45 Duplication of the gene has been suggested to contribute to ID.46 In human mesenchymal stem cells, SCRG1 expression is important for the maintenance of self-renewal and preventing osteoblastic differentiation.47 Interestingly, impaired postnatal radial glial cells self-renewal was observed in conditional Smarcc2-deficient animals,29 and abolishing the expression of Smarca4 or Smarcc2 in postnatal neuronal stem cells results in preferential onset of gliogenesis instead of neurogenesis.29, 48
Another overexpressed gene is RELN (log2FC = 3.72, padj = 3.39 × 10−4), which encodes Reelin, a large secreted matrix serine protease that is involved in layering of neurons in the cortex and cerebellum. It also regulates microtubule function in neurons and neuronal migration, and its enzymatic activity is important for the modulation of cell adhesion. Recessive RELN mutations cause lissencephaly (MIM: 257320), while dominant mutations cause epilepsy (MIM: 616436).49
Finally, CACNB4, which also has significantly increased expression (log2FC = 3.52, padj = 1.88 × 10−7), encodes a beta auxiliary subunit of voltage-gated calcium ions channels. Calcium channels play an essential role in the nervous system for neurotransmitter release as well as the regulation of gene expression.50, 51 Interestingly, CACNB4 was shown to translocate to the nucleus to regulate gene expression.52, 53 Different isoforms of the β-subunit of voltage-gated calcium ions channels have been shown to control the transcription of genes by recruiting proteins involved in DNA remodeling such as the heterochromatin protein 1.54 Also, the β-subunit was reported to downregulate Wnt signaling, a crucial player in neural development,55 through interaction with the transcription factor TCF4 which is associated with Pitt-Hopkins syndrome (MIM: 610954).53 Mutations in other subunits of voltage-gated channels have been associated with mental disorders, autism spectrum disorders (ASD), and cardiovascular problems.56, 57, 58, 59 In humans, CACNB4 mutations have been associated with episodic ataxia (MIM: 613855) and juvenile myoclonic and idiopathic generalized epilepsy (MIM: 607682).
To validate the relevance of our results and to understand the low overlap between DEGs in the two assessed individuals, we compared our results to other public datasets involving BAF-complex components. We looked at datasets in both human and mouse cell lines and primary cells in culture (Figure S2).1, 60, 61, 62, 63 The number of DEGs varies significantly between assays from just 18 to 3,038. We calculated and plotted the overlapping genes for each dataset, showing varying degrees of overlap (0% to 83%) suggesting a great heterogeneity in genes affected by BAF-complex defects, in both a subunit- and a cell type-specific manner. Thus, we hypothesize that pathogenic mutations in different domains of the same subunit, as seen in the two individuals assessed in our study, could lead to different gene expression patterns as they could affect the interaction with unique subunits or transcription factors and could explain the low overlap we obtained. Still, 48% of the DEGs in SMARCC2 individuals’ fibroblasts are found in at least one other dataset, suggesting some commonalities, and confirming a general BAF-complex-linked transcriptomic profile.
Spatiotemporal regulation of the different subunits assembly and the activity of the BAF complex are well orchestrated and essential to enable normal development and proper functioning of the nervous system.2, 64, 65 With the exception of impaired cognitive function observed in Smarcc2-deficient mice, no mutations in SMARCC2 have been associated with a particular phenotype/disease. Here we show that mutations in SMARCC2 affect gene expression, and in particular a subset of putative genes potentially implicated in neurogenesis and proper functioning of the nervous system, namely H19, SCRG1, RELN, and CACNB4. Our results further suggest that mutations of SMARCC2 may regulate pathways important for postnatal gliogenesis, differentiation, and function of astrocytes and oligodendrocytes. Further studies need to be undertaken in other models to determine the role of these genes in individuals affected by SMARCC2 mutations.
Neurodevelopmental disorders associated with pathogenic variants in BAF chromatin remodeling complex subunits present overlapping clinical phenotype. Comparing the clinical presentation of the different associated conditions (Table 23, 4, 23, 66, 67, 68, 69) emphasizes the wide clinical spectrum of the disorders and the importance of trio WES in the diagnostic process. Other than our observation that missense and splicing variants in the highly conserved SANT and SWIRM domains uniformly lead to a severe phenotype, we could not point out a clear genotype-phenotype correlation. Missense variants outside of these two domains were found mostly but not exclusively in individuals with mild presentation, suggesting variable degree of effect on BAF-complex function for missense variants. Pathogenic variants reported in SMARC subunits (SMARCA2, SMARCA4, SMARCB1, and SMARCE1) are mostly missense and in-frame deletions (Table 2) with dominant-negative effect mechanism causing Coffin-Siris and CS-like syndromes.8, 25 Recently, mutations identified in BAF-subunit DPF2 were suggested to cause CSS-like phenotype in a dominant-negative mechanism related both to missense and splicing/truncating variants through NMD escape.17
Table 2.
Summary of Clinical Presentation and Molecular Variants Associated with Coffin-Siris and Coffin-Siris-like Syndromes
| ARID1A | ARID1B | ARID2 | SMARCA2 | SMARCA4 | SMARCB1 | SMARCE1 | SOX11 | ACTL6A | DPF2 | SMARCC2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Associated condition (MIM#) | CSS 2 (614607) | CSS 1 (135900) | CSS 6 (617808) | NCBRS (601358) | CSS 4 (614609) | CSS 3 (614608) | CSS 5 (616938) | mental retardation, AD 27 (615866) | ID syndrome | CSS 7 (618027) | CSS-like |
| OMIM gene | 603024 | 614556 | 609539 | 600014 | 603254 | 601607 | 603111 | 600898 | 604958 | 601671 | 601734 |
| References | Kosho et al.;4 Kosho et al.;66 Santen et al.3 | Santen et al.3 | Gazdagh et al.67 | Santen et al.3 | Kosho et al.;4 Kosho et al.;66 Santen et al.3 | Kosho et al.;4 Kosho et al.;66 | Kosho et al.;4 Kosho et al.;66 Zarate et al.68 | Khan et al.69 | Marom et al.23 | Vasiliou et al.17 | |
| Pathogenic Variants Reported | |||||||||||
| missense | – | – | – | 4/4 | 10/12 | 4/13 | 6/6 | 4/13 | 2/3 | 5/8 | 8/15 |
| in-frame deletion | – | – | – | – | 2/12 | 9/13 (all p.Lys364del) | – | – | – | – | 1/15 |
| splice-site variants | – | 2/29 | – | – | – | – | – | – | 1/3 | 1/8 | 4/15 |
| frameshift and stop gained/nonsense | 8/8 | 25/29 | 11/14 | – | – | – | – | 2/13 | – | 2/8 | 2/15 |
| partial/full gene deletions | – | 2/29 | 3/14 | – | – | – | – | 7/13 | – | – | – |
| Clinical Presentation | |||||||||||
| Short stature | 3/8 | 3/27 | 10/14 | 1/4 | 9/12 | 11/11 | 3/5 | 4/11 | 0/3 | 4/8 | 6/13 |
| Poor weight gain | 3/6 | 11/28 | n/r | 2/4 | 4/12 | 8/9 | 4/6 | n/r | 0/3 | 4/7 | 5/13 |
| Microcephaly | 2/8 | 0/27 | 0/14 | 1/4 | 10/11 | 9/10 | 4/6 | 3/13 | n/r | 0/7 | 6/13 |
| DD and/or ID | 7/7 | 28/28 | 14/14 | 4/4 | 11/11 | 11/11 | 6/6 | 10/13 | 3/3 | 8/8 | 15/15 |
| Speech delay (first word after 12 months) | 6/6 | 28/28 | 11/11 | 4/4 | 11/11 | 10/10 | 2/6 | 11/13 | 3/3 | 8/8 | 13/15 |
| Severe speech delay (1st word ≥5 years)/no language | 5/6 | 17/28 | 5/11 | 2/4 | 6/11 | 9/10 | n/r | n/r | 0/3 | n/r | 8/15 |
| Seizures | 2/7 | 5/20 | 3/4 | 0/4 | 2/12 | 8/10 | 2/6 | 4/14 | 0/3 | n/r | 4/15 |
| Hypotonia | 7/8 | 23/27 | 2/14 | 2/4 | 8/11 | 8/11 | 2/6 | 3/13 | 0/3 | 4/7 | 13/15 |
| Brain MRI abnormalities | 7/8 | 9/25 | 6/6 | 0/3 | 6/7 | 9/9 | 3/4 | 2/3 | n/r | 3/3 | 6/12 |
| Behavioral problems: reported frequency and phenotype | 3/5; AX, HA, OCB | n/r | 10/14; ADHD, AG, AX, HA, HS (to loud noises), RG, SLD, STM | n/r | 7/8; HA, HS, IM, OCB, SAS, SI | 4/8; HA, IM, SI, TAN | 1/1; HA | 6/13; ASD, ADHD, AG, HA, SI, SLD | 2/3; ADD/ADHD, AG, IM, TAN, SLD | 3/8; STM, TAN, OCB, HA, SLD, RG, DSI/ASD | 10/15; ADHD, AG, ASD, AX, DSI, HA, HP, HS, OCB, RG, SI, SLD, TAN |
| Dysmorphic Features | |||||||||||
| reported as coarse | n/r | n/r | 9/14 | n/r | n/r | n/r | 6/6 | n/r | 1/3 | 2/8 | n/r |
| thick eyebrows | 6/8 | 24/27 | 2/7 | 4/4 | 9/12 | 11/11 | 5/6 | 1/13 | 1/3 | n/r | 6/15 |
| thick lower vermilion | 6/7 | 22/24 | 1/7 | 2/3 | 10/12 | 8/11 | 6/6 | 7/13 | n/r | n/r | 5/15 |
| hypertrichosis | 7/7 | 26/28 | 0/7 | 4/4 | 12/12 | 8/11 | n/r | 2/13 | n/r | n/r | 6/15 |
| sparse scalp hair | 3/5 | 18/28 | 0/7 | 4/4 | 5/12 | 10/11 | 5/6 | 4/13 | n/r | 6/7 | 4/15 |
| Cleft palate | 2/6 | n/r | 1/14 | n/r | 5/12 | 2/13 | 2/6 | 1/13 | 0/3 | n/r | 0/15 |
| Hand, Foot & Digital Anomalies | |||||||||||
| hypoplasia or absence of 5th distal phalanx | 6/7 | 5/22 | 2/10 | 0/3 | 10/12 | 8/11 | 2/6 | n/r | 0/3 | 0/8 | 0/15 |
| hypoplastic nails | 6/8 | 19/28 | 8/12 | 0/2 | 10/12 | 11/11 | 6/6 | 7/14 | 1/3 | 8/8 | 3/15 |
| brachydactyly | 0/8 | 11/24 | 1/10 | 1/3 | n/r | n/r | 0/6 | 0/14 | 1/3 | 5/8 | 0/15 |
| clinodactyly | n/r | n/r | 0/7 | n/r | n/r | n/r | 0/6 | 12/14 | 2/3 | 3/8 | 1/15 |
| syndactyly | n/r | n/r | 0/7 | n/r | n/r | n/r | 0/6 | 4/14 | 1/3 | n/r | 0/15 |
| Other skeletal anomalies | 2/7 | n/r | 4/14 | n/r | 1/10 | 7/9 | 3/6 | 4/14 | 1/3 | 3/8 (craniosyn-ostosis) | 7/15 |
| Laryngotracheal anomalies | 1/4 | 2/28 | 2/14 | n/r | 2/4 | n/r | 1/6 | n/r | 1/3 | n/r | 2/15 |
| Hearing loss | 2/6 | 5/22 | n/r | 0/3 | 4/12 | 8/11 | 2/6 | 2/14 | n/r | 4/8 | 2/15 |
| Cryptorchidism | 2/5 | n/r | n/r | n/r | 3/9 | 2/5 | 1/4 | 3/4 | 0/3 | n/r | 2/12 |
| Heart defect | 3/8 | n/r | 1/14 | n/r | 5/12 | 5/11 | 4/5 | n/r | 1/3 | 4/8 | 2/11 |
| Dextrocardia | 0/8 | 0/27 | n/r | 0/4 | 0/12 | 2/11 | 1/5 | n/r | 0/3 | n/r | 0/11 |
Abbreviations: AD, autosomal dominant; ADHD, attention deficit hyperactivity disorder; AG, aggressiveness; ASD, autism spectrum disorder; AX, anxiety; CCS, Coffin-Siris syndrome; DD, developmental delay; DSI, difficulties with social interactions (not qualify for ASD diagnosis); HA, hyperactivity; HP, hyperphagia; HS, hypersensitive; ID, intellectual disability; IM, impulsive; MRI, magnetic resonance imaging; NCBRS, Nicolaides-Baraitser syndrome; n/r, not reported; OCB, obsessive-compulsive behavior; OFC, occipital frontal circumference; RG, rigidity/“routine driven”; SAS, short attention span; SD, standard deviation; SI, self-injury; SLD, sleep disturbance; STM, stereotypic movements; TAN, tantrums. For comparison of clinical presentation of other BAF-related genes (including PHF6, ADNP, and TBC1D24), see Kosho et al.66
Given that SMARCC1 and SMARCC2 are paralogous genes, that they can form heterodimers or homodimers,70 and that both share functional scaffolding properties,28 it is conceivable that SMARCC1 could at least partially compensate for the loss of SMARCC2 leading to a milder phenotype in case subjects with truncating mutations in the SMARCC N-terminal region. Nevertheless, our results also suggest that the mutant mRNA of individual 4 (c.1311−3C>G) undergoes NMD as one of the consequences of the splice site mutation, thereby indicating that haploinsufficiency can also be seen in individuals with a severe phenotype. As neither RNA nor cells were available for these individuals with N-terminal truncating mutation, we cannot exclude the possibility that the mRNA could escape NMD and the truncated protein remained functional to perform essential function or to allow for a compensatory mechanism to take place. Finally, different SMARCC2 isoforms could be expressed in different tissues or at different times during development, so the location of truncating mutations could affect the phenotype depending on the affected isoform.71 The wide phenotypic variety and the lack of clear genotype-phenotype correlation of the conditions associated with BAF-complex subunits were also suggested to be attributed to a possible dosage-dependent manner of the ATP-dependent chromatin remodeling machinery.25 This, with the significant intolerance of the BAF-complex subunits genes to LoF variants, might support a contribution of LoF variants to the phenotype.
The SMARCC2-related condition presented here overlaps with other BAFopathies suggesting a CSS and Nicolaides-Baraitser-like syndrome characterized by intellectual disability, developmental delay with significant speech delay, and behavioral abnormalities. We provide evidence of dysregulated expression of H19, SCRG1, RELN, and CACNB4, the relevance of which will need to be explored in future model organism studies.
Consortia
The members of the Undiagnosed Diseases Network are as follows: David R. Adams, Mercedes E. Alejandro, Patrick Allard, Mahshid S. Azamian, Carlos A. Bacino, Ashok Balasubramanyam, Hayk Barseghyan, Gabriel F. Batzli, Alan H. Beggs, Babak Behnam, Anna Bican, David P. Bick, Camille L. Birch, Devon Bonner, Braden E. Boone, Bret L. Bostwick, Lauren C. Briere, Donna M. Brown, Matthew Brush, Elizabeth A. Burke, Lindsay C. Burrage, Shan Chen, Gary D. Clark, Terra R. Coakley, Joy D. Cogan, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Precilla D’Souza, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Ani Dillon, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Daniel C. Dorset, Emilie D. Douine, David D. Draper, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Ascia Eskin, Cecilia Esteves, Tyra Estwick, Carlos Ferreira, Brent L. Fogel, Noah D. Friedman, William A. Gahl, Emily Glanton, Rena A. Godfrey, David B. Goldstein, Sarah E. Gould, Jean-Philippe F. Gourdine, Catherine A. Groden, Andrea L. Gropman, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Lori H. Handley, Matthew R. Herzog, Ingrid A. Holm, Jason Hom, Ellen M. Howerton, Yong Huang, Howard J. Jacob, Mahim Jain, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Isaac S. Kohane, Donna M. Krasnewich, Elizabeth L. Krieg, Joel B. Krier, Seema R. Lalani, C. Christopher Lau, Jozef Lazar, Brendan H. Lee, Hane Lee, Shawn E. Levy, Richard A. Lewis, Sharyn A. Lincoln, Allen Lipson, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Marta M. Majcherska, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Ronit Marom, Julian A. Martínez-Agosto, Shruti Marwaha, Thomas May, Allyn McConkie-Rosell, Colleen E. McCormack, Alexa T. McCray, Matthew Might, Paolo M. Moretti, Marie Morimoto, John J. Mulvihill, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Neil H. Parker, Loren D.M. Pena, John A. Phillips III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Chloe M. Reuter, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Jacinda B. Sampson, Susan L. Samson, Kelly Schoch, Molly C. Schroeder, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Rebecca Signer, Edwin K. Silverman, Janet S. Sinsheimer, Kevin S. Smith, Rebecca C. Spillmann, Kimberly Splinter, Joan M. Stoler, Nicholas Stong, Jennifer A. Sullivan, David A. Sweetser, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Tiina K. Urv, Zaheer M. Valivullah, Eric Vilain, Tiphanie P. Vogel, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Patricia A. Ward, Katrina M. Waters, Monte Westerfield, Anastasia L. Wise, Lynne A. Wolfe, Elizabeth A. Worthey, Shinya Yamamoto, Yaping Yang, Guoyun Yu, Diane B. Zastrow, and Allison Zheng.
Declaration of Interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing performed at Baylor Genetics. Megan Cho and Heather McLaughlin are employees of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc. All other authors declare no competing interests.
Acknowledgments
We thank the affected individuals and their families for their participation in this study. We thank Xinyu Cao from Duke for technical support for the mRNA expression analysis. We thank the CIHR and Fonds de recherche du Québec – Santé (FRQS), Canada for clinician-scientist awards to P.M.C. Spanish individuals’ study was supported by grant PI14/00350 (Instituto de Salud Carlos III -Acción Estratégica en Salud 2013–2016; FEDER -Fondo Europeo de Desarrollo Regional). Individual 10 was evaluated through the Duke Genome Sequencing Clinic, supported by the Duke University Health system, and partially funded by UCB Celltech. Partial research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under award numbers U01HG007672 (Duke University) and U01HG007942 (Baylor College of Medicine-Sequencing). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Published: December 20, 2018
Footnotes
Supplemental Data include five tables, two figures, and Supplemental Material and Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.11.007.
Contributor Information
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Undiagnosed Diseases Network:
David R. Adams, Mercedes E. Alejandro, Patrick Allard, Mahshid S. Azamian, Carlos A. Bacino, Ashok Balasubramanyam, Hayk Barseghyan, Gabriel F. Batzli, Alan H. Beggs, Babak Behnam, Anna Bican, David P. Bick, Camille L. Birch, Devon Bonner, Braden E. Boone, Bret L. Bostwick, Lauren C. Briere, Donna M. Brown, Matthew Brush, Elizabeth A. Burke, Lindsay C. Burrage, Shan Chen, Gary D. Clark, Terra R. Coakley, Joy D. Cogan, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Precilla D’Souza, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Ani Dillon, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Daniel C. Dorset, Emilie D. Douine, David D. Draper, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Ascia Eskin, Cecilia Esteves, Tyra Estwick, Carlos Ferreira, Brent L. Fogel, Noah D. Friedman, William A. Gahl, Emily Glanton, Rena A. Godfrey, David B. Goldstein, Sarah E. Gould, Jean-Philippe F. Gourdine, Catherine A. Groden, Andrea L. Gropman, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Lori H. Handley, Matthew R. Herzog, Ingrid A. Holm, Jason Hom, Ellen M. Howerton, Yong Huang, Howard J. Jacob, Mahim Jain, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Isaac S. Kohane, Donna M. Krasnewich, Elizabeth L. Krieg, Joel B. Krier, Seema R. Lalani, C. Christopher Lau, Jozef Lazar, Brendan H. Lee, Hane Lee, Shawn E. Levy, Richard A. Lewis, Sharyn A. Lincoln, Allen Lipson, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Marta M. Majcherska, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Ronit Marom, Julian A. Martínez-Agosto, Shruti Marwaha, Thomas May, Allyn McConkie-Rosell, Colleen E. McCormack, Alexa T. McCray, Matthew Might, Paolo M. Moretti, Marie Morimoto, John J. Mulvihill, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Neil H. Parker, Loren D.M. Pena, John A. Phillips, III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Chloe M. Reuter, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Jacinda B. Sampson, Susan L. Samson, Kelly Schoch, Molly C. Schroeder, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Rebecca Signer, Edwin K. Silverman, Janet S. Sinsheimer, Kevin S. Smith, Rebecca C. Spillmann, Kimberly Splinter, Joan M. Stoler, Nicholas Stong, Jennifer A. Sullivan, David A. Sweetser, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Tiina K. Urv, Zaheer M. Valivullah, Eric Vilain, Tiphanie P. Vogel, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Patricia A. Ward, Katrina M. Waters, Monte Westerfield, Anastasia L. Wise, Lynne A. Wolfe, Elizabeth A. Worthey, Shinya Yamamoto, Yaping Yang, Guoyun Yu, Diane B. Zastrow, and Allison Zheng
Accession Numbers
Variants have been deposited in ClinVar under submission number SUB4878282.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
GeneBank, https://www.ncbi.nlm.nih.gov/genbank/
GeneMatcher, https://www.genematcher.org/
Genetic Intolerance, http://genic-intolerance.org/
Genome Browser, https://genome.ucsc.edu/
GnomAD Browser, http://gnomad.broadinstitute.org/
Human Phenotype Ontology (HPO), http://human-phenotype-ontology.org/
Human Splicing Finder, http://www.umd.be/HSF3/
Missense Tolerance Ratio (MTR) Tool, http://mtr-viewer.mdhs.unimelb.edu.au/mtr-viewer/
OMIM, https://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2
PROVEAN, http://provean.jcvi.org/
Supplemental Data
References
- 1.Barutcu A.R., Lajoie B.R., Fritz A.J., McCord R.P., Nickerson J.A., van Wijnen A.J., Lian J.B., Stein J.L., Dekker J., Stein G.S., Imbalzano A.N. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016;26:1188–1201. doi: 10.1101/gr.201624.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronan J.L., Wu W., Crabtree G.R. From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santen G.W., Aten E., Vulto-van Silfhout A.T., Pottinger C., van Bon B.W., van Minderhout I.J., Snowdowne R., van der Lans C.A., Boogaard M., Linssen M.M., Coffin-Siris consortium Coffin-Siris syndrome and the BAF complex: genotype-phenotype study in 63 patients. Hum. Mutat. 2013;34:1519–1528. doi: 10.1002/humu.22394. [DOI] [PubMed] [Google Scholar]
- 4.Kosho T., Okamoto N., Coffin-Siris Syndrome International Collaborators Genotype-phenotype correlation of Coffin-Siris syndrome caused by mutations in SMARCB1, SMARCA4, SMARCE1, and ARID1A. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:262–275. doi: 10.1002/ajmg.c.31407. [DOI] [PubMed] [Google Scholar]
- 5.Sousa S.B., Hennekam R.C., Nicolaides-Baraitser Syndrome International Consortium Phenotype and genotype in Nicolaides-Baraitser syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:302–314. doi: 10.1002/ajmg.c.31409. [DOI] [PubMed] [Google Scholar]
- 6.Hoyer J., Ekici A.B., Endele S., Popp B., Zweier C., Wiesener A., Wohlleber E., Dufke A., Rossier E., Petsch C. Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am. J. Hum. Genet. 2012;90:565–572. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halgren C., Kjaergaard S., Bak M., Hansen C., El-Schich Z., Anderson C.M., Henriksen K.F., Hjalgrim H., Kirchhoff M., Bijlsma E.K. Corpus callosum abnormalities, intellectual disability, speech impairment, and autism in patients with haploinsufficiency of ARID1B. Clin. Genet. 2012;82:248–255. doi: 10.1111/j.1399-0004.2011.01755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Houdt J.K., Nowakowska B.A., Sousa S.B., van Schaik B.D., Seuntjens E., Avonce N., Sifrim A., Abdul-Rahman O.A., van den Boogaard M.J., Bottani A. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat. Genet. 2012;44:445–449. doi: 10.1038/ng.1105. S441. [DOI] [PubMed] [Google Scholar]
- 9.Vandeweyer G., Helsmoortel C., Van Dijck A., Vulto-van Silfhout A.T., Coe B.P., Bernier R., Gerdts J., Rooms L., van den Ende J., Bakshi M. The transcriptional regulator ADNP links the BAF (SWI/SNF) complexes with autism. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:315–326. doi: 10.1002/ajmg.c.31413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santen G.W., Kriek M., van Attikum H. SWI/SNF complex in disorder: SWItching from malignancies to intellectual disability. Epigenetics. 2012;7:1219–1224. doi: 10.4161/epi.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neale B.M., Kou Y., Liu L., Ma’ayan A., Samocha K.E., Sabo A., Lin C.F., Stevens C., Wang L.S., Makarov V. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loe-Mie Y., Lepagnol-Bestel A.M., Maussion G., Doron-Faigenboim A., Imbeaud S., Delacroix H., Aggerbeck L., Pupko T., Gorwood P., Simonneau M., Moalic J.M. SMARCA2 and other genome-wide supported schizophrenia-associated genes: regulation by REST/NRSF, network organization and primate-specific evolution. Hum. Mol. Genet. 2010;19:2841–2857. doi: 10.1093/hmg/ddq184. [DOI] [PubMed] [Google Scholar]
- 13.Chesi A., Staahl B.T., Jovičić A., Couthouis J., Fasolino M., Raphael A.R., Yamazaki T., Elias L., Polak M., Kelly C. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrier S.A., Bodurtha J.N., Burton B., Chudley A.E., Chiong M.A., D’avanzo M.G., Lynch S.A., Musio A., Nyazov D.M., Sanchez-Lara P.A. The Coffin-Siris syndrome: a proposed diagnostic approach and assessment of 15 overlapping cases. Am. J. Med. Genet. A. 2012;158A:1865–1876. doi: 10.1002/ajmg.a.35415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsurusaki Y., Okamoto N., Ohashi H., Kosho T., Imai Y., Hibi-Ko Y., Kaname T., Naritomi K., Kawame H., Wakui K. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 16.Bramswig N.C., Caluseriu O., Lüdecke H.J., Bolduc F.V., Noel N.C., Wieland T., Surowy H.M., Christen H.J., Engels H., Strom T.M., Wieczorek D. Heterozygosity for ARID2 loss-of-function mutations in individuals with a Coffin-Siris syndrome-like phenotype. Hum. Genet. 2017;136:297–305. doi: 10.1007/s00439-017-1757-z. [DOI] [PubMed] [Google Scholar]
- 17.Vasileiou G., Vergarajauregui S., Endele S., Popp B., Büttner C., Ekici A.B., Gerard M., Bramswig N.C., Albrecht B., Clayton-Smith J., Deciphering Developmental Disorders Study Mutations in the BAF-complex subunit DPF2 are associated with Coffin-Siris syndrome. Am. J. Hum. Genet. 2018;102:468–479. doi: 10.1016/j.ajhg.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hempel A., Pagnamenta A.T., Blyth M., Mansour S., McConnell V., Kou I., Ikegawa S., Tsurusaki Y., Matsumoto N., Lo-Castro A., DDD Collaboration Deletions and de novo mutations of SOX11 are associated with a neurodevelopmental disorder with features of Coffin-Siris syndrome. J. Med. Genet. 2016;53:152–162. doi: 10.1136/jmedgenet-2015-103393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zweier C., Rittinger O., Bader I., Berland S., Cole T., Degenhardt F., Di Donato N., Graul-Neumann L., Hoyer J., Lynch S.A. Females with de novo aberrations in PHF6: clinical overlap of Borjeson-Forssman-Lehmann with Coffin-Siris syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:290–301. doi: 10.1002/ajmg.c.31408. [DOI] [PubMed] [Google Scholar]
- 20.Nicolaides P., Baraitser M. An unusual syndrome with mental retardation and sparse hair. Clin. Dysmorphol. 1993;2:232–236. [PubMed] [Google Scholar]
- 21.Pretegiani E., Mari F., Renieri A., Penco S., Dotti M.T. Nicolaides-Baraitser syndrome: defining a phenotype. J. Neurol. 2016;263:1659–1660. doi: 10.1007/s00415-016-8194-0. [DOI] [PubMed] [Google Scholar]
- 22.Campeau P.M., Hennekam R.C., DOORS syndrome collaborative group DOORS syndrome: phenotype, genotype and comparison with Coffin-Siris syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:327–332. doi: 10.1002/ajmg.c.31412. [DOI] [PubMed] [Google Scholar]
- 23.Marom R., Jain M., Burrage L.C., Song I.W., Graham B.H., Brown C.W., Stevens S.J.C., Stegmann A.P.A., Gunter A.T., Kaplan J.D. Heterozygous variants in ACTL6A, encoding a component of the BAF complex, are associated with intellectual disability. Hum. Mutat. 2017;38:1365–1371. doi: 10.1002/humu.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helsmoortel C., Vulto-van Silfhout A.T., Coe B.P., Vandeweyer G., Rooms L., van den Ende J., Schuurs-Hoeijmakers J.H., Marcelis C.L., Willemsen M.H., Vissers L.E. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bögershausen N., Wollnik B. Mutational landscapes and phenotypic spectrum of SWI/SNF-related intellectual disability disorders. Front. Mol. Neurosci. 2018;11:252. doi: 10.3389/fnmol.2018.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ring H.Z., Vameghi-Meyers V., Wang W., Crabtree G.R., Francke U. Five SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin (SMARC) genes are dispersed in the human genome. Genomics. 1998;51:140–143. doi: 10.1006/geno.1998.5343. [DOI] [PubMed] [Google Scholar]
- 27.Tuoc T.C., Boretius S., Sansom S.N., Pitulescu M.E., Frahm J., Livesey F.J., Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev. Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Narayanan R., Pirouz M., Kerimoglu C., Pham L., Wagener R.J., Kiszka K.A., Rosenbusch J., Seong R.H., Kessel M., Fischer A. Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 2015;13:1842–1854. doi: 10.1016/j.celrep.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 29.Tuoc T., Dere E., Radyushkin K., Pham L., Nguyen H., Tonchev A.B., Sun G., Ronnenberg A., Shi Y., Staiger J.F. Ablation of BAF170 in developing and postnatal dentate gyrus affects neural stem cell proliferation, differentiation, and learning. Mol. Neurobiol. 2017;54:4618–4635. doi: 10.1007/s12035-016-9948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben-David E., Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol. Psychiatry. 2013;18:1054–1056. doi: 10.1038/mp.2012.148. [DOI] [PubMed] [Google Scholar]
- 31.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez F., Caro-Llopis A., Roselló M., Oltra S., Mayo S., Monfort S., Orellana C. High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J. Med. Genet. 2017;54:87–92. doi: 10.1136/jmedgenet-2016-103964. [DOI] [PubMed] [Google Scholar]
- 33.Zhu X., Petrovski S., Xie P., Ruzzo E.K., Lu Y.F., McSweeney K.M., Ben-Zeev B., Nissenkorn A., Anikster Y., Oz-Levi D. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet. Med. 2015;17:774–781. doi: 10.1038/gim.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinodoz M., Royer-Bertrand B., Cisarova K., Di Gioia S.A., Superti-Furga A., Rivolta C. DOMINO: using machine learning to predict genes associated with dominant disorders. Am. J. Hum. Genet. 2017;101:623–629. doi: 10.1016/j.ajhg.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrovski S., Gussow A.B., Wang Q., Halvorsen M., Han Y., Weir W.H., Allen A.S., Goldstein D.B. The intolerance of regulatory sequence to genetic variation predicts gene dosage sensitivity. PLoS Genet. 2015;11:e1005492. doi: 10.1371/journal.pgen.1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aravind L., Iyer L.M. The SWIRM domain: a conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol 3. 2002 doi: 10.1186/gb-2002-3-8-research0039. RESEARCH0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoneyama M., Tochio N., Umehara T., Koshiba S., Inoue M., Yabuki T., Aoki M., Seki E., Matsuda T., Watanabe S. Structural and functional differences of SWIRM domain subtypes. J. Mol. Biol. 2007;369:222–238. doi: 10.1016/j.jmb.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 40.Boyer L.A., Latek R.R., Peterson C.L. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 41.Lelieveld S.H., Wiel L., Venselaar H., Pfundt R., Vriend G., Veltman J.A., Brunner H.G., Vissers L.E.L.M., Gilissen C. Spatial clustering of de novo missense mutations identifies candidate neurodevelopmental disorder-associated genes. Am. J. Hum. Genet. 2017;101:478–484. doi: 10.1016/j.ajhg.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabory A., Jammes H., Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 43.Raveh E., Matouk I.J., Gilon M., Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol. Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han C.L., Ge M., Liu Y.P., Zhao X.M., Wang K.L., Chen N., Meng W.J., Hu W., Zhang J.G., Li L., Meng F.G. LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J. Neuroinflammation. 2018;15:103. doi: 10.1186/s12974-018-1139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dron M., Dandoy-Dron F., Guillo F., Benboudjema L., Hauw J.J., Lebon P., Dormont D., Tovey M.G. Characterization of the human analogue of a Scrapie-responsive gene. J. Biol. Chem. 1998;273:18015–18018. doi: 10.1074/jbc.273.29.18015. [DOI] [PubMed] [Google Scholar]
- 46.Kashevarova A.A., Nazarenko L.P., Skryabin N.A., Salyukova O.A., Chechetkina N.N., Tolmacheva E.N., Sazhenova E.A., Magini P., Graziano C., Romeo G. Array CGH analysis of a cohort of Russian patients with intellectual disability. Gene. 2014;536:145–150. doi: 10.1016/j.gene.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 47.Aomatsu E., Takahashi N., Sawada S., Okubo N., Hasegawa T., Taira M., Miura H., Ishisaki A., Chosa N. Novel SCRG1/BST1 axis regulates self-renewal, migration, and osteogenic differentiation potential in mesenchymal stem cells. Sci. Rep. 2014;4:3652. doi: 10.1038/srep03652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ninkovic J., Steiner-Mezzadri A., Jawerka M., Akinci U., Masserdotti G., Petricca S., Fischer J., von Holst A., Beckers J., Lie C.D. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wasser C.R., Herz J. Reelin: neurodevelopmental architect and homeostatic regulator of excitatory synapses. J. Biol. Chem. 2017;292:1330–1338. doi: 10.1074/jbc.R116.766782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mochida S. Presynaptic calcium channels. Neurosci. Res. 2018;127:33–44. doi: 10.1016/j.neures.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 51.Greer P.L., Greenberg M.E. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Ronjat M., Kiyonaka S., Barbado M., De Waard M., Mori Y. Nuclear life of the voltage-gated Cacnb4 subunit and its role in gene transcription regulation. Channels (Austin) 2013;7:119–125. doi: 10.4161/chan.23895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rima M., Daghsni M., Lopez A., Fajloun Z., Lefrancois L., Dunach M., Mori Y., Merle P., Brusés J.L., De Waard M., Ronjat M. Down-regulation of the Wnt/β-catenin signaling pathway by Cacnb4. Mol. Biol. Cell. 2017;28:3699–3708. doi: 10.1091/mbc.E17-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tadmouri A., Kiyonaka S., Barbado M., Rousset M., Fablet K., Sawamura S., Bahembera E., Pernet-Gallay K., Arnoult C., Miki T. Cacnb4 directly couples electrical activity to gene expression, a process defective in juvenile epilepsy. EMBO J. 2012;31:3730–3744. doi: 10.1038/emboj.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mulligan K.A., Cheyette B.N. Wnt signaling in vertebrate neural development and function. J. Neuroimmune Pharmacol. 2012;7:774–787. doi: 10.1007/s11481-012-9404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinggera A., Striessnig J. Cav 1.3 (CACNA1D) L-type Ca2+ channel dysfunction in CNS disorders. J. Physiol. 2016;594:5839–5849. doi: 10.1113/JP270672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zamponi G.W. Targeting voltage-gated calcium channels in neurological and psychiatric diseases. Nat. Rev. Drug Discov. 2016;15:19–34. doi: 10.1038/nrd.2015.5. [DOI] [PubMed] [Google Scholar]
- 58.Breitenkamp A.F., Matthes J., Herzig S. Voltage-gated calcium channels and autism spectrum disorders. Curr. Mol. Pharmacol. 2015;8:123–132. doi: 10.2174/1874467208666150507105235. [DOI] [PubMed] [Google Scholar]
- 59.Soldatov N.M. CACNB2: an emerging pharmacological target for hypertension, heart failure, arrhythmia and mental disorders. Curr. Mol. Pharmacol. 2015;8:32–42. doi: 10.2174/1874467208666150507093258. [DOI] [PubMed] [Google Scholar]
- 60.Wang X., Lee R.S., Alver B.H., Haswell J.R., Wang S., Mieczkowski J., Drier Y., Gillespie S.M., Archer T.C., Wu J.N. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 2017;49:289–295. doi: 10.1038/ng.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raab J.R., Runge J.S., Spear C.C., Magnuson T. Co-regulation of transcription by BRG1 and BRM, two mutually exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin. 2017;10:62. doi: 10.1186/s13072-017-0167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hodges H.C., Stanton B.Z., Cermakova K., Chang C.Y., Miller E.L., Kirkland J.G., Ku W.L., Veverka V., Zhao K., Crabtree G.R. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol. 2018;25:61–72. doi: 10.1038/s41594-017-0007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen H., Kerimoglu C., Pirouz M., Pham L., Kiszka K.A., Sokpor G., Sakib M.S., Rosenbusch J., Teichmann U., Seong R.H. Epigenetic regulation by BAF complexes limits neural stem cell proliferation by suppressing Wnt signaling in late embryonic development. Stem Cell Reports. 2018;10:1734–1750. doi: 10.1016/j.stemcr.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sokpor G., Xie Y., Rosenbusch J., Tuoc T. Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front. Mol. Neurosci. 2017;10:243. doi: 10.3389/fnmol.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Son E.Y., Crabtree G.R. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:333–349. doi: 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kosho T., Miyake N., Carey J.C. Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: historical review and recent advances using next generation sequencing. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:241–251. doi: 10.1002/ajmg.c.31415. [DOI] [PubMed] [Google Scholar]
- 67.Gazdagh G., Blyth M., Scurr I., Turnpenny P.D., Mehta S.G., Armstrong R., McEntagart M., Newbury-Ecob R., Tobias E.S., Study D.D.D. Extending the clinical and genetic spectrum of ARID2 related intellectual disability. A case series of 7 patients. Eur. J. Med. Genet. 2018 doi: 10.1016/j.ejmg.2018.04.014. Published online April 23, 2018. pii: S1769-7212(18)30016-8. [DOI] [PubMed] [Google Scholar]
- 68.Zarate Y.A., Bhoj E., Kaylor J., Li D., Tsurusaki Y., Miyake N., Matsumoto N., Phadke S., Escobar L., Irani A. SMARCE1, a rare cause of Coffin-Siris Syndrome: Clinical description of three additional cases. Am. J. Med. Genet. A. 2016;170:1967–1973. doi: 10.1002/ajmg.a.37722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khan U., DDD Study. Baker E., Clayton-Smith J. Observation of cleft palate in an individual with SOX11 mutation: indication of a role for SOX11 in human palatogenesis. Cleft Palate Craniofac. J. 2018;55:456–461. doi: 10.1177/1055665617739312. [DOI] [PubMed] [Google Scholar]
- 70.Wang W., Xue Y., Zhou S., Kuo A., Cairns B.R., Crabtree G.R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 71.Kazantseva A., Sepp M., Kazantseva J., Sadam H., Pruunsild P., Timmusk T., Neuman T., Palm K. N-terminally truncated BAF57 isoforms contribute to the diversity of SWI/SNF complexes in neurons. J. Neurochem. 2009;109:807–818. doi: 10.1111/j.1471-4159.2009.06005.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.