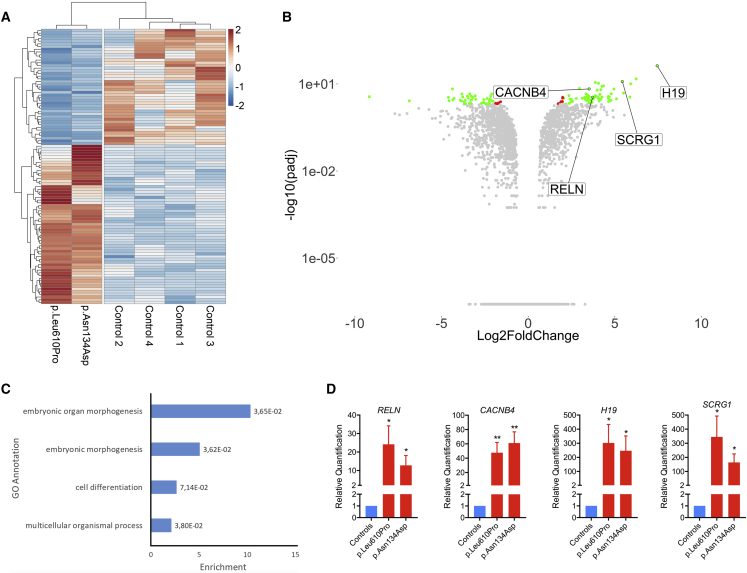
Figure 4.
Fibroblasts Harboring p.Leu610Pro or p.Asn134Asp SMARCC2 Mutation Have Differential Gene Expression Patterns When Compared to Fibroblasts from Healthy Control Subjects
RNA-seq was performed on fibroblasts from four healthy controls and two affected individuals to assay gene expression.
(A) Heatmap of differentially expressed genes (DEGs). Log2 fold change (Log2FC) and p values adjusted for 10% false discovery rate (padj) were calculated for the two individuals and four control subjects, respectively pooled together using the “DESeq2” R package. Normalized count values from “DESeq2” were plotted and scaled row-wise using the “pheatmap” R package for genes with a padj lower than 0.01 and an absolute Log2FC value higher than 2.
(B) Volcano plot showing differentially regulated genes as fold-change versus adjusted p values. Log2FC and –log10(padj) values were plotted using the “ggplot2” R package as a volcano plot. Red dots represent genes with a padj lower than 0.01 and a abs(Log2FC) lower than 2. Green dots represent genes with a padj lower than 0.01 and a abs(log2FC) higher than 2.
(C) Histogram of the enriched GO annotations with FDR adjusted p values lower than 0.01 calculated using the GOrilla web application using DEGs for the two individuals pooled together.
(D) RT-qPCR analysis of mRNA expression profile for RELN, CACNB4, H19, and SCRG1 with p.Leu610Pro and p.Asn134Asp SMARCC2 mutations. The data are presented as “relative quantification” mean ± SEM from 6 independent control subjects (N = 2). A Student’s t test was used to determine the statistical significance. ∗p ≤ 0.05 versus control group; ∗∗p ≤ 0.01 versus control group.