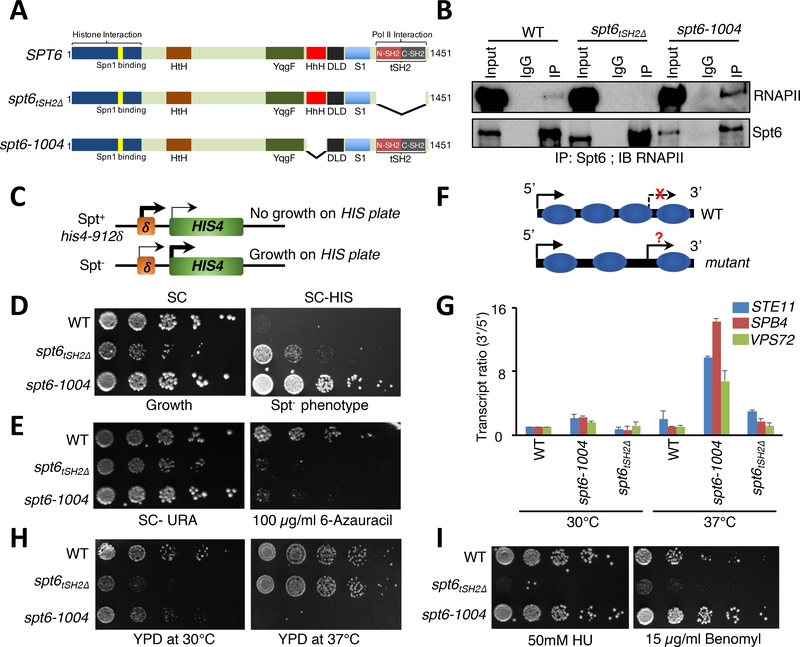
Figure 1. Uncoupling Spt6-RNAPII interaction reveals distinct Spt6 phenotypes.
(A) Domain organization and forms of Spt6 used in this study. HtH (Helix-turn-Helix domain); YqgF (RNase H-like domain); HhH (Helix-hairpin-Helix domain); S1 (S1 domain); DLD (Death-like-Domain); and tSH2 (Tandem Src2 homology domain). (B) Spt6 interacts with RNAPII via a tSH2 domain. Co-immunoprecipitation experiments were performed for Spt6 and RNAPII, and interaction was assessed by immunoblot analysis with various phospho-Ser2-CTD specific antibodies. (C) Schematic of the Spt- phenotype. Insertion of a Ty element at the HIS4 locus suppresses expression. Spt- mutants allow expression of HIS4 and growth on medium lacking histidine. (D) Spt- phenotypic assay. 5-fold serial dilutions of the indicated strains were spotted on synthetic complete (SC) and SC medium lacking histidine. (E) Spt6 is required for transcription elongation. Spotting assays showing the sensitivity of spt6 mutants on 6-azauracil plates. (F) Schematic illustrating normal and cryptic (intragenic) transcription. In wild-type (WT) cells, intragenic sites of transcription are normally suppressed but can be activated in mutants that perturb chromatin integrity. (G) qRT-PCR assay to validate the generation of intragenic sense transcripts using primers spanning the 5’ and the 3’ regions of STE11, SPB4, and VPS72 genes in WT, spt6–1004, and spt6tSH2Δ mutant cells. (H) and (I) Spotting assays showing functionally distinct phenotypes of the spt6–1004 and spt6tSH2Δ mutants upon exposure to heat and genotoxic agents, respectively. 5-fold serial dilutions were spotted on the indicated medium. Plates were incubated at 30°C or 37°C and photographs were taken after 4 days.