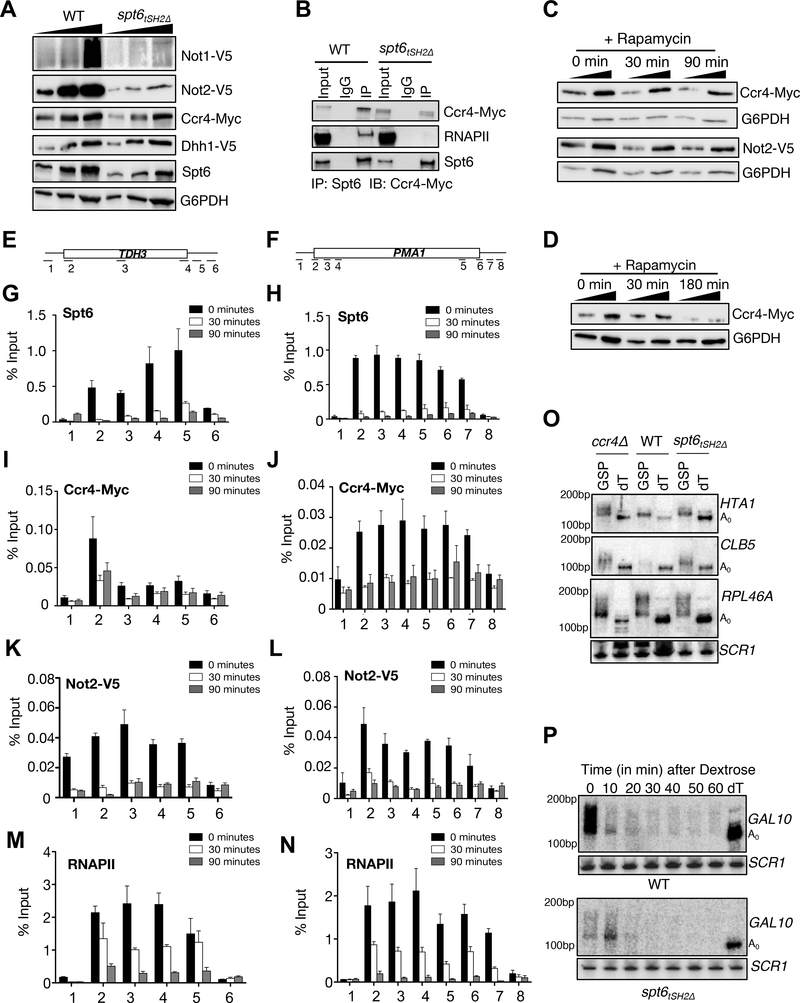
Figure 6. Spt6 recruits the Ccr4-Not complex to chromatin.
Interaction of Spt6 with RNAPII is required for optimal levels of Ccr4-Not complex members. (A) Exponentially growing WT and spt6tSH2Δ mutant cells were subjected to immunoblot analysis (after TCA lysis) to detect changes in the levels of Ccr4-Not complex members (B) Co-immunoprecipitation showing an interaction between Spt6 and Ccr4 in WT and the spt6tSH2Δ mutant. (C) and (D) Immunoblots showing changes in the levels of Ccr4 and Not2 with Spt6 anchor-away at 30, 90 minutes and 3h, respectively. (E) and (F) Schematic representation of the primer locations across PMA1 and TDH3 genes. (G) and (H) ChIP-qPCR of Spt6 kinetics on PMA1 and TDH3 genes following rapamycin treatment. ChIP-qPCR showing changes in the occupancy of Ccr4 (I and J) and Not2 (K and L) and RNAPII (M and N) on PMA1 and TDH3 genes respectively, following Spt6 depletion. While Spt6 and Ccr4/Not2 are no longer detectable at the 30 minute time-point, we note that RNAPII is still detected, thereby suggesting a role for Spt6 in recruitment of Ccr4-Not (O) Spt6 is required for proper mRNA metabolism and poly(A) tail length. Total RNA was isolated using acid phenol method and 10μg was digested with RNAse H in the presence of gene specific primer (GSP) and/or oligo-dT12–16 (dT); A0 represents the completely deadenylated RNA species. Use of the GSP alone gives the length of the poly(A) tail whereas use of the GSP with dT allows for the determination of A0. Total RNA was separated in urea-PAGE, blotted on to membranes and probed with oligo probes. (P) High resolution PAGE Northern blot showing the rate of deadenylation of GAL10 mRNA.