Fig. 4.
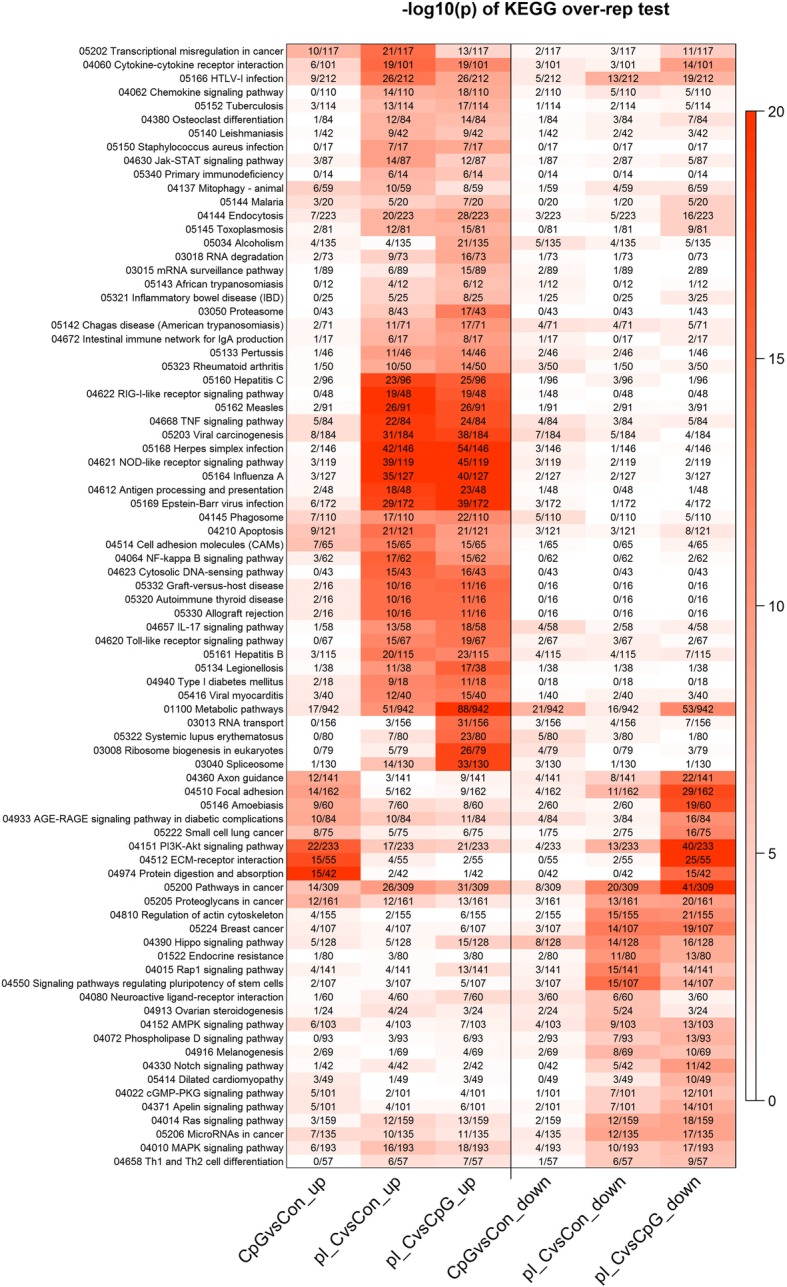
Representation of KEGG pathways. Each pathway is a row and each gene set is a column. The red color intensity shows the –log10 (raw p-value), i.e., a value of 6 = 1e-6. As the traditional FDR correction is not appropriate for KEGG or GO testing, we used an extremely low raw p-value to focus on the most highly significant pathways or terms. The numbers in the boxes show the number of genes in the pathway that are significantly up or downregulated, divided by the total number of genes in the pathway. CpG DNAvsCon_up = upregulated genes in the CpG DNA vs control comparison; pI_CvsCon_up = upregulated genes in the poly(I:C) vs control comparison; pI_CvsCpG DNA_up = upregulated genes in the poly(I:C) vs CpG DNA comparison; CpG DNAvsCon_down = downregulated genes in the CpG DNA vs control comparison; pI_CvsCon_down = downregulated genes in the poly(I:C) vs control comparison; pI_CvsCpG DNA_down = downregulated genes in the poly(I:C) vs CpG DNA comparison
