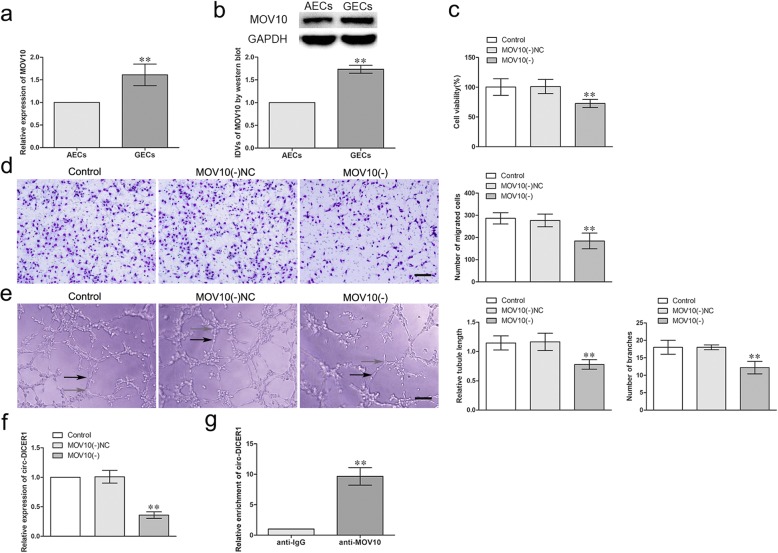
Fig. 1.
The expressions of MOV10 in GECs and knockdown of MOV10 suppressed the viability, migration and tube formation of GECs in vitro and MOV10 combined with circ-DICER1 and regulated its expression. a-b The mRNA and protein expressions of MOV10 in AECs and GECs were evaluated by qRT-PCR and western blot. GAPDH was used as an endogenous control. IDVs represents “Integrated Density Values”. Data represent means ± SD (n = 5, each group). **P < 0.01 vs. AECs group. c Effects of MOV10 knockdown on cell viability of GECs were detected by CCK-8 assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs MOV10 (−) NC group. d Effects of MOV10 knockdown on migration of GECs were detected by Transwell assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs MOV10 (−) NC group. Scale bar represents 30 μm. e Effects of MOV10 knockdown on tube formation of GECs were measured by Matrigel tube formation assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs MOV10 (−) NC group. Scale bar represents 30 μm. f Effects of MOV10 knockdown on expression of circ-DICER1 were detected by qRT-PCR. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs MOV10 (−) NC group. g Relative enrichment of circ-DICER1 in anti-IgG and anti-MOV10 were detected by RNA immunoprecipitation assay. Data represent means ± SD (n = 3, each group). **P < 0.01 vs. anti-IgG group