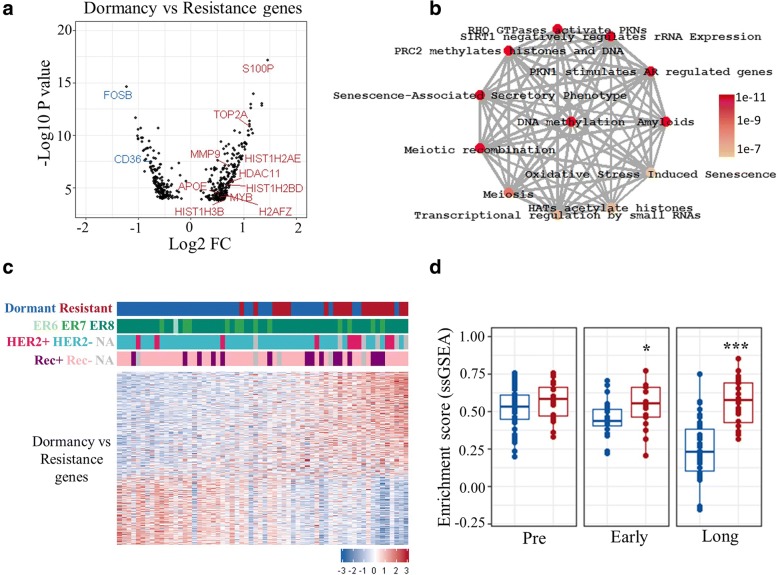
Fig. 4.
Comparative analysis of dormant and resistant tumours. a Volcano plot showing differentially expressed genes between long-term treated dormant and resistant tumours (dormancy versus resistance genes). Some upregulated and downregulated genes in resistant tumours are highlighted in red and blue, respectively. b Significantly enriched pathways for dormancy versus resistance genes (p < 0.01; ReactomePA). Grey edges connecting the nodes indicates genes shared between the nodes/pathways, and the width of the edge is scaled by the number of common genes. Colours indicates the significance (p value) where red is a lower p value. c Heatmap showing partial separation of long-term treated dormant and resistant samples using dormancy versus resistance genes (a total of 419; 170 downregulated and 249 upregulated genes). Colours are log2 mean-centred values with red indicating high and blue indicating low expression. Genes are sorted by fold-change (FC) values from most to least up/downregulated. Samples are sorted by sum expression of upregulated genes. ER6, ER7, and ER8 correspond to oestrogen receptor (ER) Allred scores 6, 7, and 8, respectively. d Comparison of single sample gene enrichment analysis (ssGSEA) scores of dormancy versus resistance upregulated genes between dormant and acquired resistant tumours. Dormant (blue); resistant (red); before (pre, ≤ 0 days), early-on (early, 13–120 days) and long-term (long, > 120 days) neoadjuvant aromatase inhibitor therapy with letrozole. ***p < 0.001; *p < 0.05. NA not available, Rec+ recurrence, Rec– recurrence free