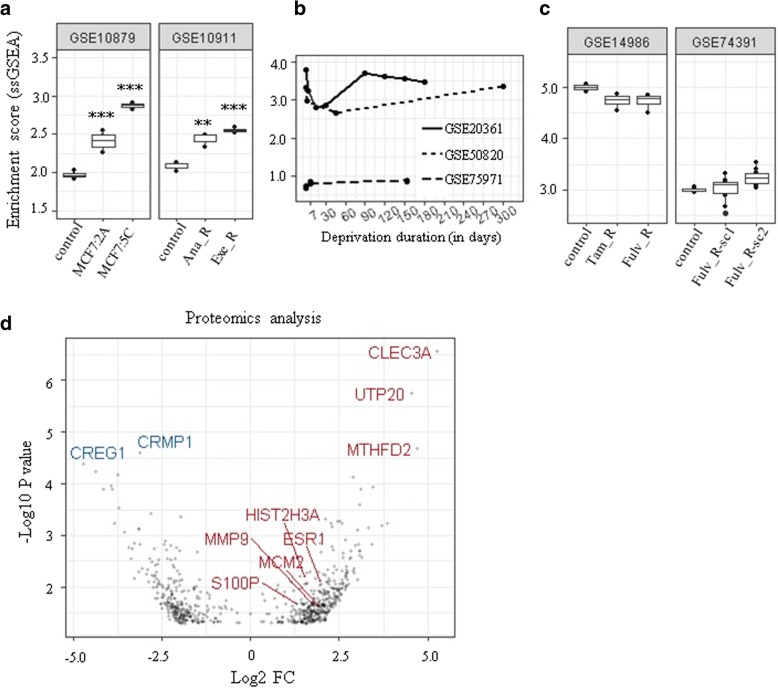
Fig. 5.
Validation of results using in-vitro gene expression data from resistant cell lines and proteomics analysis. a Normalised enrichment scores of differently upregulated genes (a total of 249) between long-term treated dormant and resistant tumours calculated using single sample gene set enrichment analysis (ssGSEA) in aromatase inhibitor-resistant cells. Scores were significantly higher (**p < 0.01, ***p < 0.001) in two aromatase inhibitor-resistant cell lines, MCF7:2A and MCF7:5C, which were clonally derived from MCF7 breast cancer cells following long-term oestrogen deprivation (LTED) compared with control/sensitive MCF7 cells (n = 4). Anastrozole-resistant (Ana_R) and exemestane-resistant (Exe_R) MCF7aro cells had significantly higher scores compared with control (n = 3). b Dynamic changes in enrichment scores of LTED MCF7 cells in three different datasets. c Scores in tamoxifen-resistant (Tam_R) and fulvestrant-resistant (Fulv_R) and drug-sensitive (control) MCF7 cells (n = 4, n = 10). d Volcano plot showing differentially expressed proteins between long-term treated dormant and resistant tumours (p < 0.05). Some overlapping features between transcriptomics and proteomics analysis and the most upregulated and downregulated proteins are highlighted in red and blue, respectively. FC fold-change, sc subclone