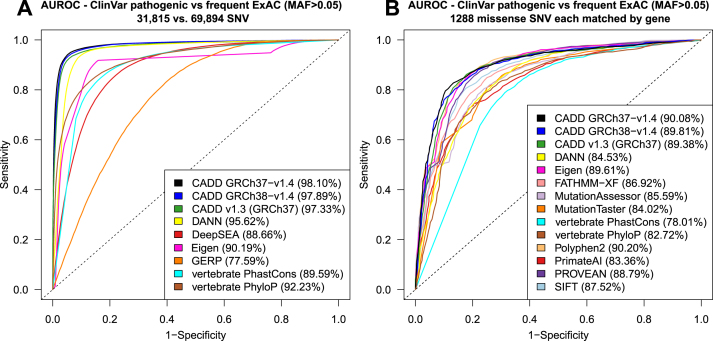
Figure 2.
Performance of CADD in comparison to other scores. Different scores are compared by area under the receiver operating characteristic (AUROC) in terms of how well they separate known pathogenic variants (ClinVar pathogenic) from frequent exome variants (ExAC, mean allele frequency >5%, assumed to be neutral): (A) All variants of the two sets, and (B) missense variants only, with matching genes between the two sets. PolyPhen2 and PROVEAN, two dedicated protein missense variant scores, perform on par with CADD and Eigen, while all other scores have a lower AUROC. The performance of CADD GRCh38-v1.4 is not significantly different from the other CADD releases. The results for more missense scores and non-coding variants are shown in Supplementary Figure S1.