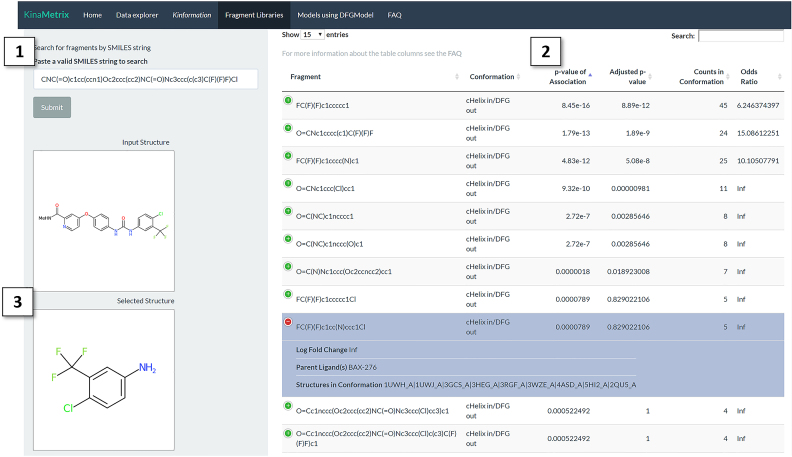
Figure 3.
Identifying chemical substructures that confer conformational specificity. Under the ‘Fragment Libraries’ panel and the ‘Search by SMILES’ tab users can search the substructure library on KinaMetrix using a SMILES string. For example, the drug sorafenib was searched against our substructure library. (1) The input SMILES string search box. (2) This table displays the matched substructures to the input chemical. These substructures are annotated with their association for a kinase conformation, as well as information about their parent ligand and their co-crystalized kinase structures. The top substructure matches that are associated with the αC-helix in/DFG out (CIDO) conformational state are shown. (3) The 2D-representation of both the input SMILES string and the selected substructure from the table.