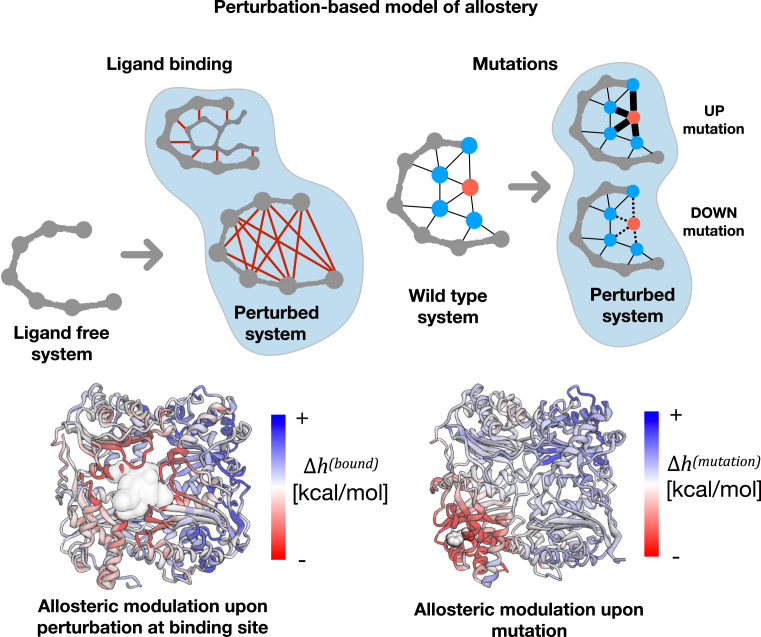
Figure 1.
Perturbation-based model of allostery used in calculations. Top row: schemes of implementing the perturbations; ligand binding is mimicked by introducing strong interactions between residues of the binding site; mutations are modeled by increasing/decreasing force constants for interactions of mutated residue with its neighbors. Bottom row: Examples of the visualization of allosteric modulation caused by the ligand binding (left) and mutation (right).