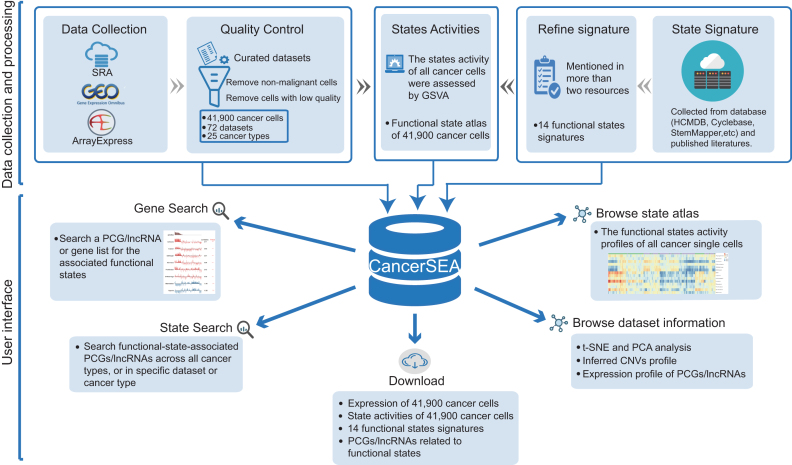
Figure 1.
Overview of CancerSEA database. All scRNAseq datasets were collected from SRA, GEO and ArrayExpress, and were manually annotated and curated. For quality control, we removed non-malignant single cells and cells with low quality. In addition, we collected and refined 14 functional states signatures. State activities of cancer cells were assessed by GSVA. All data resource were deposited in CancerSEA, and displayed in web pages (gene search, state search, browse, download).